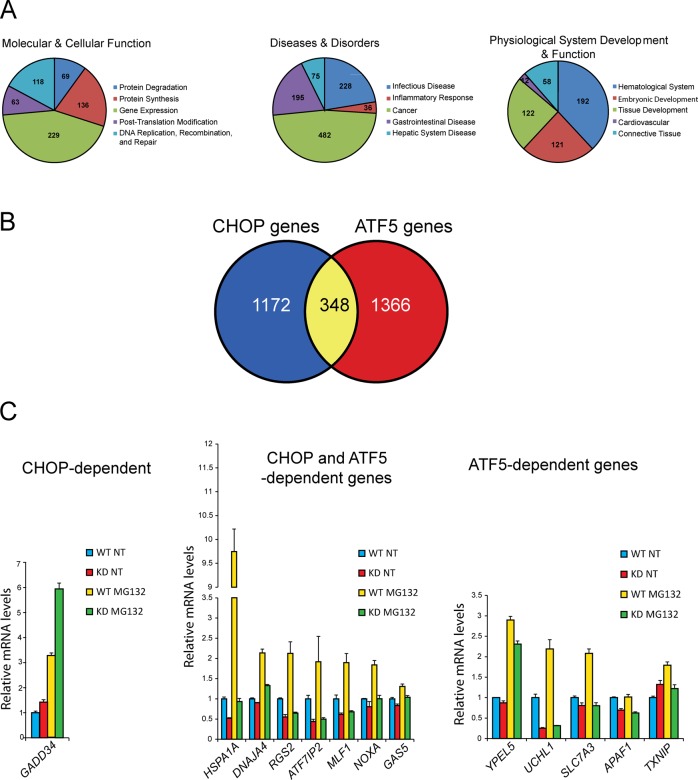
FIGURE 6:
Genome-wide analysis of gene expression indicates that ATF5 is required for activation of proapoptotic target genes. Results from a microarray analysis of WT and ATF5-KD2 cells treated with MG132 or no stress indicated those genes that significantly required ATF5 for full induction in response to proteasome inhibition. These ATF5-target genes were then analyzed for functional groupings and compared with those genes determined in our microarray analysis to be statistically dependent on CHOP for full induction in response to MG132 treatment. (A) Pie charts generated by IPA analysis, highlighting functional classes of ATF5-dependent genes. (B) Venn diagram indicating the number of genes that required only CHOP for full induction response to MG132 treatment, those genes dependent on both CHOP and ATF5, and genes dependent on only ATF5. (C) Comparative bar graph showing that GADD34 required only CHOP for full induction in response to proteasome inhibition, examples of genes whose induction showed a statistically significant dependence on both CHOP and ATF5, and those genes requiring only ATF5; values represent mean fluorescence intensity (MFI) in WT and ATF5-KD2 cells upon MG132 treatment or no treatment (NT).