Figure 1.
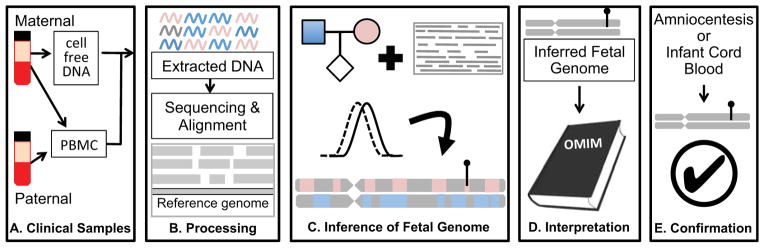
Overview of noninvasive fetal whole genome sequencing. (a). Sample collection. Parental blood samples are collected in the first or second trimester. After centrifugation, parental DNA is extracted from peripheral blood mononuclear cells (PBMC) or buffy coat, while cfDNA is isolated from the maternal plasma. (b). Sample processing. Extracted DNA is amplified for library preparation and sequenced to high depth. Reads are aligned to a reference genome to identify variant alleles carried by one or both parents. (c). Inference of fetal genome. A statistical model combines known parental genotypes and alleles observed in cfDNA reads to predict fetal inheritance. High-impact mutations, whether inherited or de novo, are identified (lollypop). (d). Interpretation. Identified variants are compared to catalogs of known disease-associated mutations. (e). Confirmation. A subset of clinically actionable predicted mutations is confirmed with conventional procedures such as amniocentesis. Accuracy of genome inference can be assessed post-hoc with DNA extracted from cord blood after delivery.
