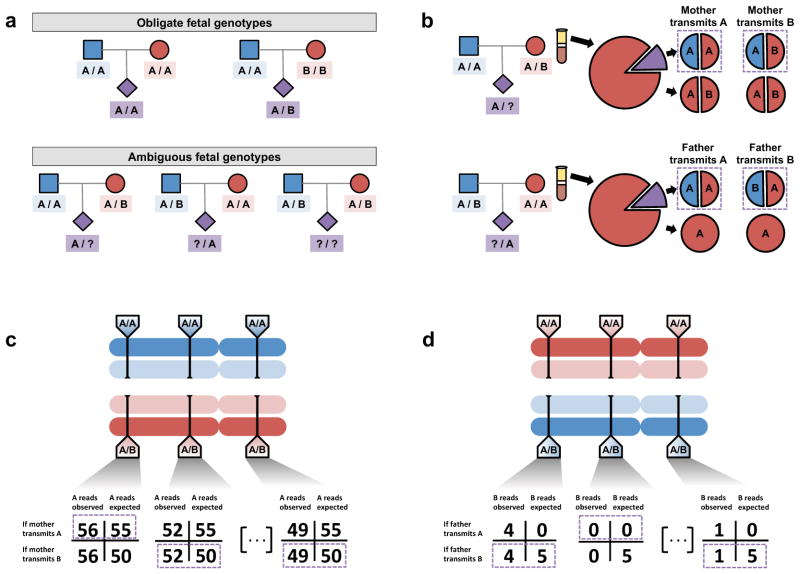
Figure 2.
Inference of the fetal genome on a site-by-site basis. (a). Observed parental genotypes at a given site constrain the possible fetal genotypes. At the vast majority of sites, both parents are homozygotes and the fetal genotype is unambiguous. (b). Expected cfDNA makeup in the maternal plasma at maternal (top) and paternal (bottom) heterozygous sites. At these sites, either allele could be transmitted, yielding different expected allelic fractions. (c). Schematic of inference of fetal inheritance at maternal heterozygous sites. Numbers shown assume a constant sequencing depth of 100X at each site and do not represent real data. After sequencing the cfDNA, observed allele counts are compared to expected allele counts at each site, and in each case, the more likely scenario is chosen (purple boxes). (d). Schematic of inference of fetal inheritance at paternal heterozygous sites. Numbers are presented as in (c). At each site, presence of the B allele is unexpected in cases where the father transmits the “A” allele. At the rightmost site, the single observed “B” allele could be evidence of true “B” transmission or an error introduced during the sequencing process.