Table 1. Evaluation results on T-Cell signaling.
| LP model | LP model REP | DEPN | Sachs et al. | random | |
| TP | 3 | 9 | 2 | 12 | 2.72 |
| TN | 95 | 102 | 97 | 99 | 91.72 |
| FP | 9 | 2 | 7 | 5 | 13.28 |
| FN | 14 | 8 | 15 | 5 | 13.28 |
| SP | 0.91** | 0.98** | 0.93** | 0.95** | 0.86 |
| SN | 0.18 | 0.53** | 0.12 | 0.71** | 0.16 |
| PR | 0.25 | 0.82** | 0.22 | 0.71** | 0.16 |
| AC | 0.81* | 0.92** | 0.82* | 0.92** | 0.76 |
The table shows performance measures for the network inference on flow cytometry data regarding signaling downstream of CD3, CD28 and LFA-1 in CD4 T-cells. Network inference was performed using the linear program (LP), Deterministic Effects Propagation Networks (DEPN), random guessing, and a Bayesian network model as implemented by Sachs et
al. TP = true positives, TN = true negatives, FP = false positives, SP = specificity, SN = sensitivity, PR = precision, AC = accuracy. The column “LP model REP” corresponds to the evaluation results of the LP model where the reversely inferred edges and reported indirect regulations are counted as true positives. Statistically significant differences are marked with
T-cells. Network inference was performed using the linear program (LP), Deterministic Effects Propagation Networks (DEPN), random guessing, and a Bayesian network model as implemented by Sachs et
al. TP = true positives, TN = true negatives, FP = false positives, SP = specificity, SN = sensitivity, PR = precision, AC = accuracy. The column “LP model REP” corresponds to the evaluation results of the LP model where the reversely inferred edges and reported indirect regulations are counted as true positives. Statistically significant differences are marked with  (
(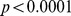 ) and
) and  (
(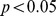 ), respectively.
), respectively.
