Table 2. Evaluation results on ErbB signaling.
| LP model | DEPN | random | |
| TP | 9 | 6 | 14.72 |
| TN | 71 | 71 | 58.72 |
| FP | 15 | 15 | 27.28 |
| FN | 33 | 36 | 27.28 |
| SP | 0.83** | 0.83** | 0.68 |
| SN | 0.21** | 0.14** | 0.35 |
| PR | 0.38 | 0.28 | 0.35 |
| AC | 0.63 | 0.60 | 0.57 |
Shown are comparative performance measurements for network inference on reverse phase protein array data regarding ErbB signaling in breast cancer cells. Network inference was performed using our linear programming (LP) approach, Deterministic Effects Propagation Networks (DEPN), and random guessing of a network. Results were compared with a gold standard network from the String database. TP = true positives, TN = true negatives, FP = false positives, SP = specificity, SN = sensitivity, PR = precision, AC = accuracy. Statistically significant differences are marked with  (
(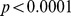 ) and
) and  (
(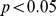 ), respectively.
), respectively.
