Table 4. Non-Embeddability – D1 Human, Rat, Mouse Triad (8394 Alignments).
| STEPS a | ||||||||
| Edge | Codon position | 1 | 2 | 3 | 4 | 5 | NEb Matrices | NE Processesc |
| Human | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0d | |
| 3 | 16 | 16 | 0 | 3 | 91 | 107 | 6 (5.6) | |
| Mouse | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| Rat | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | |
 Steps to identify Non-embeddability 1.
Steps to identify Non-embeddability 1. 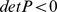 , 2. Negative eigenvalues have odd algebraic multiplicity, 3. Complex eigenvalues occur in non-conjugate pairs, 4. The set of eigenvalues,
, 2. Negative eigenvalues have odd algebraic multiplicity, 3. Complex eigenvalues occur in non-conjugate pairs, 4. The set of eigenvalues, 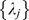 , lie outside the region in the complex plane, 5.
, lie outside the region in the complex plane, 5. 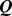 – negative off-diagonals – threshold −0.1,
– negative off-diagonals – threshold −0.1,  NE = Non-Embeddable,
NE = Non-Embeddable,  No. rejections of
No. rejections of 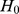 from parametric bootstrap scheme with a p-value
from parametric bootstrap scheme with a p-value 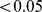 (percentage of total tests),
(percentage of total tests),  1 Alignment failed to find stable estimates.
1 Alignment failed to find stable estimates.
