Figure 5. Catalytically inactive or folate-binding deficient GNMT mutants are capable of the antiproliferative effect.
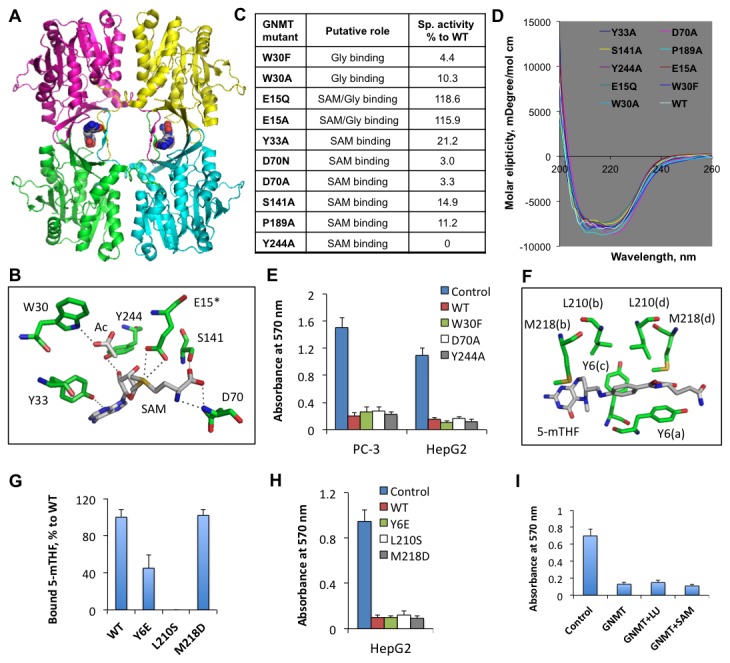
A. Crystal structure of GNMT tetramer (RCSB Protein Data Bank 3ths; subunits are shown in different colors) with bound 5-MTHF monoglutamate (two molecules shown in spacefill mode are bound per tetramer). B. Positions of amino acid residues in the GNMT catalytic center (from RCSB Protein Data Bank 1XVA). Acetate (Ac) is the competitive inhibitor of Gly and presumably occupies the same position in the active center. Glu 15 (E15*) is from a different subunit. Dotted lines indicate hydrogen bonds. C and D. The enzyme activities and CD spectra of GNMT mutants, analyzed in this study. E. The MTT proliferation assay of cells transfected with empty vector (control), wild type GNMT (WT), or corresponding mutants. Error bars represent ± S.D., n =3. F. Folate binding site at the GNMT subunit interface (as shown in panel A); Selected for mutagenesis are residues within close distance to 5-MTHF molecule (these residues are from all four subunits, which are denoted in parentheses). G. Binding of 5-MTHF by GNMT mutants. Error bars represent ± S.D., n =2. H. The MTT proliferation assay of cells transfected with empty vector (control), wild type GNMT (WT), or folate-binding deficient mutants mutants. Error bars represent ± S.D., n =3. I. The supplementation with excessively high media folate or SAM does not rescue cells from the GNMT antiproliferative effect. Error bars represent ± S.D., n =3.
