| 0 |
Set the value of unobserved mortality among LTF, UM, to zero. |
| 1 |
Randomly select a value of CD4ij to represent the median CD4 count in CD4 band j of study i, for all i and j. Imputed values are drawn from a specified beta distributions defined over the interval between the lower and upper bounds of the CD4 band (or if no upper bound, the lower bound plus 100). We use left-skewed distribution used to impute the median CD4 count in the highest (open ended) CD4 strata, right-skewed distributions for bands with 0 as a lower bound, and symmetric distributions for interior bands (shown in Appendix S3 in File S1). |
| 2 |
Randomly impute values of LTFi for two studies where LTFU was not reported. |
| 3 |
Use equation (3) to compute the value of the dependent variable, ln(TMij). |
| 4 |
Estimate equation (2) by weighted ordinary least squares of ln(TMij) on ln(CD4ij). To correct for heteroskedasticity, weight each observation by its sample size. For studies which did not report sample size by CD4 band (n = 5), randomly allocate total sample size across bands. |
| 5 |
Repeat step 1–4 1,000 times |
| 6 |
Combine the 1,000 estimated regression results to compute a pooled estimate of β, its standard error and the F-statistic measuring the goodness-of-fit of the regression. Apply Rubin’s formula[35]–[37] to estimate the pooled standard error of β:where W is the average within-imputation variance of 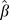 , and B is the variance of the estimator across M = 1,000 imputations. , and B is the variance of the estimator across M = 1,000 imputations. |
| 7 |
Repeat steps 1–6 10 times, incrementing the value of UM by 0.1 each time |
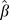 , and B is the variance of the estimator across M = 1,000 imputations.
, and B is the variance of the estimator across M = 1,000 imputations.