Abstract
Hot springs are the most common infectious source of Legionella pneumophila in Japan. However, little is known about the association between L. pneumophila and environmental waters other than hot springs. In this study, water samples from 22 environmental water sites were surveyed; of the 22 samples, five were L. pneumophila positive (23%). L. pneumophila was mainly isolated from ashiyu foot spas, a type of hot spring for the feet (3/8, 38%). These isolates had genetic loci or genes that encoded the virulence factors of L. pneumophila. Moreover, these isolates showed higher intracellular growth and stronger cytotoxicity compared with the reference strain. These results suggest that ashiyu foot spa can be the original source for L. pneumophila infection.
1. Introduction
Legionella pneumophila is the causative agent of legionellosis. In humans, L. pneumophila can induce Legionnaires' disease and Pontiac fever. Legionnaires' disease is a form of severe pneumonia, while Pontiac fever produces acute flu-like symptoms without pneumonia [1]. A number of factors including type II and type IV secretion systems, a pore-forming toxin, type IV pili, flagella, and heat shock proteins [2–7] contribute to L. pneumophila virulence. L. pneumophila is a facultative intracellular Gram-negative bacterium that can reside and multiply within free-living amoebae in environmental waters. L. pneumophila can withstand temperatures of 0–68°C and a pH range of 5.0–8.5 and survive in most environments for long periods [8]. L. pneumophila mainly lives in natural and man-made aquatic environment such as ponds, hot springs, fountains, cooling towers, and portable waters [8]. Hot springs and public baths are known to be most common source of L. pneumophila outbreaks in Japan [9–11]. Abundance information about the relationship between L. pneumophila and hot springs and public baths has been accumulated, but there is little information regarding L. pneumophila in environmental waters other than hot springs and public baths.
In this study, 22 environmental water places were surveyed in Yamaguchi Prefecture, Japan, and L. pneumophila was isolated from five sites.
2. Materials and Methods
2.1. Bacteria and Culture Conditions
Legionella pneumophila Lp02 and the ΔdotA mutant, Lp03 [2, 5], were maintained as frozen glycerol stocks and cultured on N-(2-acetamido-) 2-aminoethanesulphonic acid (ACES)-buffered charcoal-yeast extract broth containing 1.5% agar (CYET) or liquid ACES-buffered yeast extract broth (AYET) supplemented with 100 μg/mL thymidine.
Isolation of L. pneumophila was performed using CYET supplemented with glycine (Wako, Osaka, Japan, 3 mg/mL), vancomycin HCl (Wako, 1 μg/mL), polymyxin B (Sigma, MD, USA, 79.2 IU/mL), and sulfate cycloheximide (Wako, 80 μg/mL) (GVPC agar) [12]. Isolated bacteria were grown on CYET at 37°C or in AYET with shaking [13].
2.2. Specimen Collection and Preparation
Samples were collected from 22 environmental water sites. Eight samples were collected from ashiyu foot spas, seven were from water fountains, four were from basins of shrine, and three were from ponds (Table 1). Five hundred milliliters of sample was collected from each site in sterile bottles or small plastic containers and centrifuged at 3000 rpm for 20 min at 4°C. The deposits were resuspended in 500 μL distilled water as concentrates. Concentrated samples were heated at 50°C for 30 min and spread onto the surface of GVPC agar. Plates were incubated at 37°C and they were inspected daily.
Table 1.
Detection of Legionella pneumophila from environmental waters.
| Place | No. of collected points | No. of positive points | Positive rate (%) |
|---|---|---|---|
| Water fountain | 7 | 1 | 14 |
| Ashiyu foot spa | 8 | 3 | 38 |
| Basin | 4 | 0 | 0 |
| Pond | 3 | 1 | 33 |
|
| |||
| Total | 22 | 5 | 23 |
2.3. PCR Analysis
The primers used for PCR analysis are summarized in Table 2. After denaturation of the bacterial chromosomal DNA template at 95°C for 5 min, 35 cycles of PCR amplification were performed using expand high fidelity PCR system (Roche, Basel, Switzerland).
Table 2.
Oligonucleotides.
| Name/region | Sequence (5′-3′) | Reference |
|---|---|---|
| lvh1/lvhB3 | attgggagcttctggcaata | This study |
| lvh2/lvhB3 | gctggggtgacctttgaata | This study |
| rtx1/rtxA | gctgcaaccacctctttgat | This study |
| rtx2/rtxA | caggggctggttatgttgat | This study |
| dot1/dotA | caaatccggcattcaaaatc | This study |
| dot2/dotA | ctattgtcgccttgggtgtt | This study |
| hsp1/hsp60 | gcgaatcgttgttaccaaagaaaac | [15] |
| hsp2/hsp60 | caatttgacgcattggagattcaatag | [15] |
| mip1/mip | ggtgactgcggctgttatgg | [16] |
| mip2/mip | ggccaataggtccgccaacg | [16] |
2.4. Serotyping
Serotypes of isolated bacteria were determined based on their reactions during the immunoagglutination serotyping with Legionella immune sera (Denka Seiken, Tokyo, Japan).
2.5. Cell Lines and Culture Conditions
HeLa cells were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM, Sigma) containing 10% heat-inactivated fetal bovine serum (FBS, Biowest, Paris, France). A human monocytic cell line, THP-1 cells, was grown at 37°C and 5% CO2 in RPMI 1640 medium (Sigma), containing 10% heat-inactivated FBS. THP-1 cells were differentiated with 100 nM phorbol 12-myristate 13-acetate (PMA, Sigma) at 48 h prior to use.
2.6. Intracellular Invasion and Growth Assays
Bacteria were added to a monolayer of HeLa cells or THP-1 cells in 48-well tissue culture dishes at multiplicity of infection (MOI) of 100 or 1, respectively. These plates were centrifuged for 5 min at 900 ×g and incubated for 1 h at 37°C. Extracellular bacteria were killed by gentamicin (50 μg/mL) treatment for 1 h. To measure the invasion efficiency, cells were washed twice with phosphate-buffered saline (PBS) and lysed with cold distilled water. To measure the intracellular growth, the cells were incubated in fresh medium at 37°C for particular time and washed three times with PBS, followed by lysis with cold distilled water. Colony forming units (CFU) were determined by serial dilution on CYET.
2.7. Cytotoxicity Measurement
Bacteria were added to a monolayer of HeLa cells or THP-1 cells in 48-well tissue culture dishes at MOI of 100 or 1, respectively. These plates were centrifuged for 5 min at 900 ×g and incubated for 1 h at 37°C. Extracellular bacteria were killed by gentamicin (50 μg/mL) treatment for 1 h. Cells were washed twice with PBS and incubated in fresh medium at 37°C. At 24 or 48 h after incubation, the supernatants of infected cells were collected. Cytotoxicity was determined by measuring LDH release using a Cytotoxicity Detection KitPLUS (LDH) (Roche) according to the manufacturer's instructions.
2.8. Statistical Analysis
Data are expressed as the mean of triplicate samples from three identically performed experiments, and the error bars represented the standard deviations. Statistical analyses were performed using Student's t-test. Statistically significant differences are indicated by asterisks (∗, P < 0.05).
3. Results
3.1. Isolation and Identification
Twenty-two samples were collected from environmental water sites in Yamaguchi Prefecture, Japan. Samples were concentrated and spread on GVPC agar. Five possible colonies were obtained. Three were isolated from ashiyu foot spas, one was isolated from a water fountain, and the other was isolated from a pond.
To confirm whether these isolates were L. pneumophila or not, the presence of L. pneumophila specific gene, mip [14], was tested by PCR. The mip gene was detected in all isolates, indicating that these isolates were L. pneumophila. We named these isolates Twr292, Ymt294, Ofk308, Ymg289, and Bnt314 (Tables 1 and 3).
Table 3.
Isolation of Legionella pneumophila from PCR-positive sites.
| Strain | Place | CFU/100 mL | Serotype |
|---|---|---|---|
| Ymg289 | Water fountain | 1 | I |
| Twr292 | Ashiyu foot spa | 128 | I |
| Ymt294 | Ashiyu foot spa | 2 | I |
| Ofk308 | Ashiyu foot spa | 2 | IV |
| Bnt314 | Pond | 4 | IV |
The serotypes of these five isolates were then determined by immunoagglutination serotyping. Twr292, Ymt294, and Ymg289 were classified into serotype I, and Ofk308 and Bnt314 were classified into serotype IV (Table 3).
3.2. Growth in Liquid Medium
We compared the growth of the five isolates in AYET medium with that of the virulent reference strain Lp02 and the avirulent ΔdotA mutant Lp03, which lacks a functional Dot/Icm secretion system. Twr292, Ofk308, Ymg289, and Bnt314 showed comparable growth with Lp02 and Lp03. In contrast Ymt294 had shown lower growth rate. After 48 h, the number of Ymt294 was almost one-tenth of Lp02 and Lp03 (Figure 1).
Figure 1.
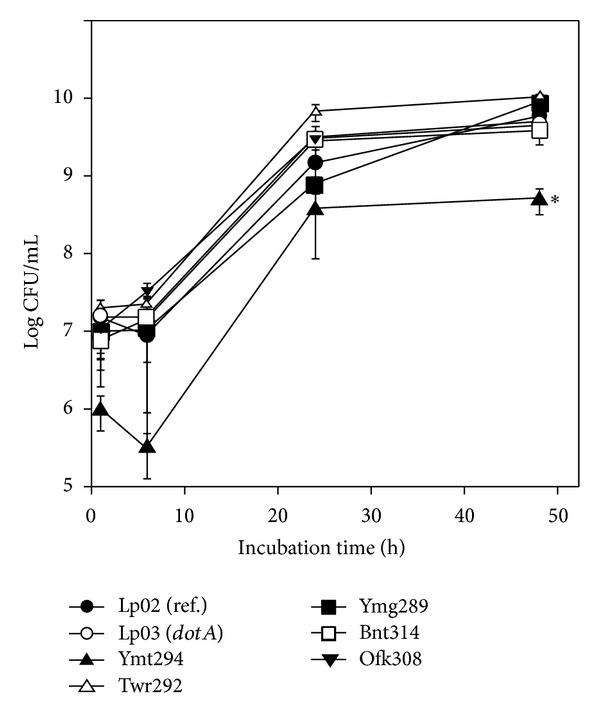
Growth of L. pneumophila isolates in liquid medium. Bacteria were grown in AYET. After 1, 24, and 48 h of incubation, samples were diluted with PBS and spread on CYET. All values represent the average and the standard deviation for three identical experiments. Statistically significant differences compared with the control are indicated by asterisks (∗, P < 0.05).
3.3. Invasion, Intracellular Growth, and Cytotoxicity in HeLa Cells
To investigate the intracellular behavior of the isolates, their invasion, growth, and cytotoxicity in HeLa cells were examined. HeLa cells were infected with the isolates, and the number of invaded L. pneumophila was counted at 1 h after infection. Ymt294, Twr292, and Ymg289 invaded HeLa cells more than ten times higher than reference strain Lp02 (Figure 2(a)).
Figure 2.
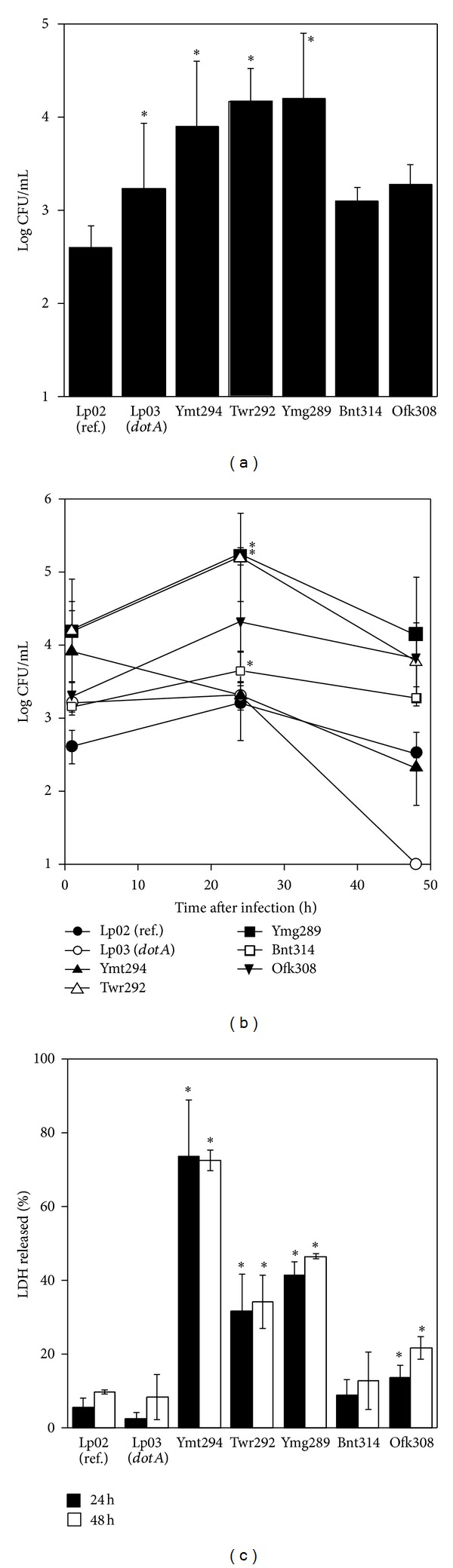
Invasion, intracellular growth, and cytotoxicity in HeLa cell. (a) HeLa cells were infected with L. pneumophila strains for 1 h. The infected cells were cultured in the presence of 50 μg/mL gentamicin. After 1 h of incubation, the infected cells were washed with PBS and lysed with cold distilled water. CFU were determined by serial dilution on CYET. (b) HeLa cells were infected with L. pneumophila strains at MOI of 100 for 1 h. The infected cells were cultured in the presence of 50 μg/mL gentamicin. The infected cells were cultured for 1, 24, and 48 h and washed with PBS followed by lysis with cold distilled water. CFU were determined by serial dilution on CYET. (c) HeLa cells were infected with L. pneumophila strains for 1 h. The infected cells were cultured in the presence of 50 μg/mL gentamicin for 1 h. After 24 or 48 h incubation, the cells were washed and cultured in fresh medium. The supernatants of infected cells were collected, and the release of LDH was measured. All values represent the average and the standard deviation for three identical experiments. Statistically significant differences compared with the control are indicated by asterisks (∗, P < 0.05).
Intracellular growth of the isolates was measured by counting intracellular bacteria numbers at 24 and 48 h after infection. At 24 h after infection, Twr292, Ymg289, and Bnt314 showed higher growth and the bacterial number was more than ten times as compared with the reference strain Lp02. At 48 h after infection, the numbers of all isolates were decreased. The ΔdotA mutant Lp03 failed to replicate in HeLa cells, as previously reported [17] (Figure 2(b)).
The cytotoxicity of isolates was measured by LDH release assay and phase-contrast microscopy. At 24 and 48 h after infection, Ymt294, Twr292, and Ymg289 showed high cytotoxicity (Figure 2(c)). At 24 h after infection with isolates, cells were damaged and detached from the culture plates (Figures 4(a)–4(c) and data not shown).
Figure 4.
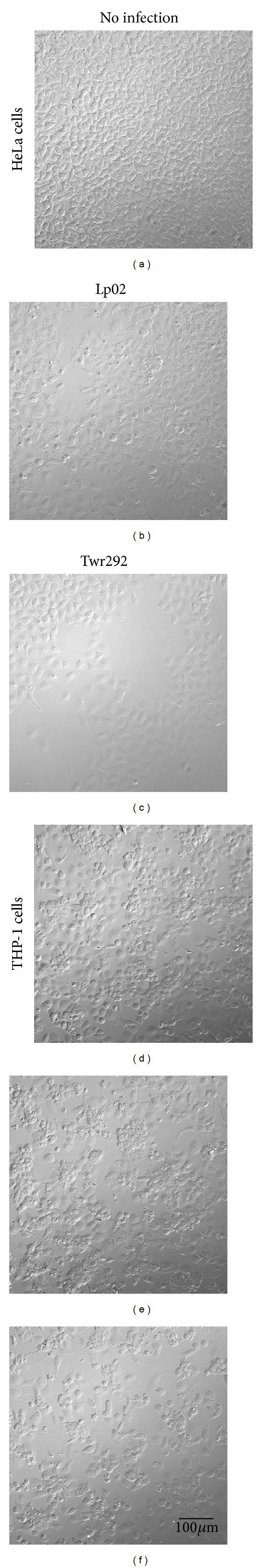
Cytotoxicity in HeLa and THP-1 cells. HeLa cells ((a)–(c)) and THP-1 cells ((d)-(e)) were infected with L. pneumophila strains Lp02 ((b) and (e)) or Twr292 ((c) and (f)) for 1 h. The infected cells were cultured in the presence of 50 μg/mL gentamicin for 1 h. The cells were washed and cultured in fresh medium. After 24 h of incubation, the condition of cells was observed using phase-contrast microscope.
3.4. Intracellular Growth and Cytotoxicity in THP-1 Cells
L. pneumophila resides predominantly in macrophages after infection; therefore, the growth and cytotoxicity of isolates were examined in a human macrophage cell line, THP-1 cells. At 24 h and 48 h after infection, all isolates showed potent growth. The numbers of these isolates were ten times higher than the reference strain Lp02. The ΔdotA mutant Lp03 failed to grow in THP-1 cells (Figure 3(a)). Moreover, all isolates showed higher cytotoxicity than the reference strain in THP-1 cells. Particularly Twr292 induced strong cytotoxicity (Figure 3(b)). Damaged and detached THP-1 cells were observed with phase-contrast microscopy after cells were infected with Twr292 (Figures 4(d)–4(f)).
Figure 3.
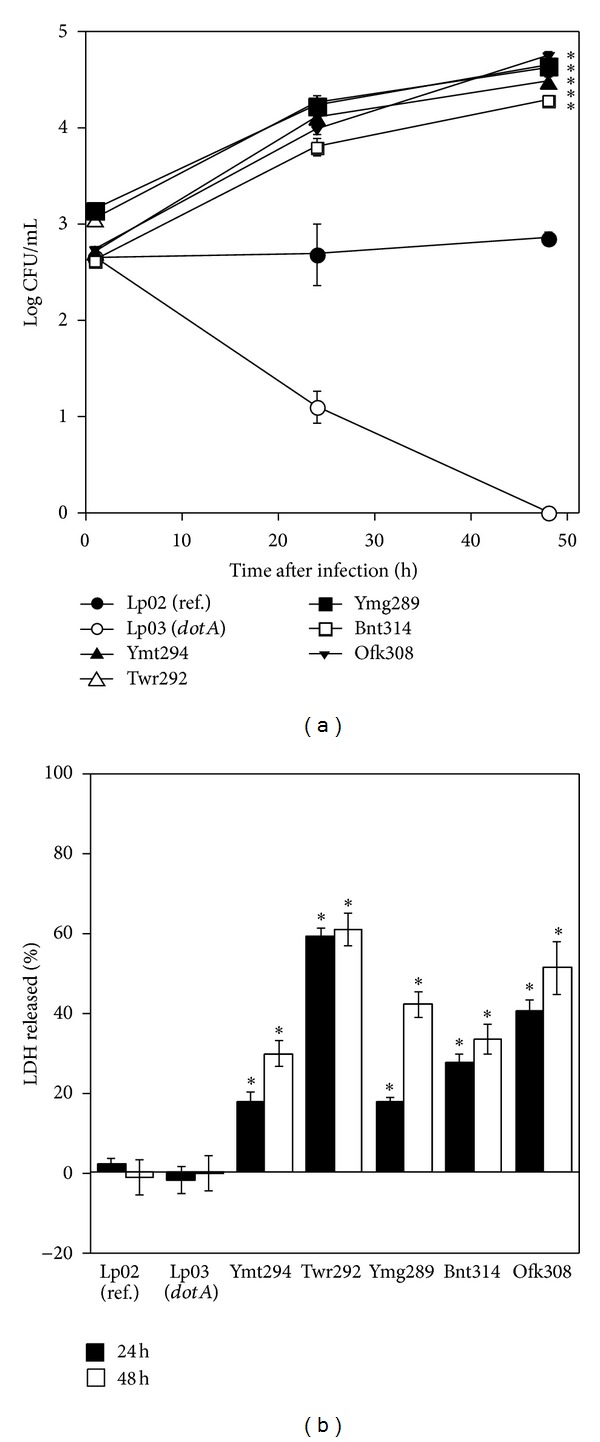
Intracellular growth and cytotoxicity in THP-1 cell. (a) THP-1 cells were infected with L. pneumophila strains at MOI of 1 for 1 h. The infected cells were cultured in the presence of 50 μg/mL gentamicin. The infected cells were cultured for 1, 24, and 48 h and washed with PBS followed by lysis with cold distilled water. CFU were determined by serial dilution on CYET. (b) THP-1 cells were infected with L. pneumophila strains for 1 h. The infected cells were cultured in the presence of 50 μg/mL gentamicin for 1 h. After 24 or 48 h of incubation, the cells were washed and cultured in fresh medium. The supernatants of infected cells were collected, and LDH release was measured. All values represent the average and the standard deviation for three identical experiments. Statistically significant differences compared with the control are indicated by asterisks (∗, P < 0.05).
3.5. Detection of Loci and Genes Related to Virulence Factor
To estimate whether these isolates are pathogenic to humans, the presence of genetic loci of dot, lvh, and rtx that encode typical virulence factors of L. pneumophila was examined. Loci of dot and lvh encode components of type IV secretion system that play an important role in intracellular growth [18]. Locus rtx encodes proteins involved in adherence, cytotoxicity, and pore formation [19]. The presence of dot, lvh, and rtx loci was tested by detecting dotA, lvhB3, and rtxA genes located in these loci, respectively, by PCR. The presence of the hsp60 gene was also examined. hsp60 encodes a 60 kDa heat shock protein (Hsp60) that enhances invasion and elicits cytokine expression in macrophages [20, 21]. These genes were detected in all five isolates (Table 4), indicating that these isolates are human pathogenic.
Table 4.
Detection of loci and genes related to virulence factor.
| Region | Lp02 | Lp03 | Ymg289 | Twr292 | Ymt294 | Ofk308 | Bnt314 |
|---|---|---|---|---|---|---|---|
| lvhB3 | + | + | + | + | + | + | + |
| rtxA | + | + | + | + | + | + | + |
| dotA | + | − | + | + | + | + | + |
| hsp60 | + | + | + | + | + | + | + |
4. Discussion
In Japan, hot springs are reported to be the major infectious source for L. pneumophila [9–11]. However, there is little information about L. pneumophila in environmental waters other than hot springs. In this study, we tested 22 environmental water sites in Yamaguchi Prefecture, Japan. Eight were from ashiyu foot spas, seven were from water fountains, four were from basins, and three were from ponds. L. pneumophila was isolated from five sites (23%) (Table 1). Three were isolated from ashiyu foot spas (38%), one was isolated from a water fountain (14%), and the other was isolated from pond (33%). Interestingly, L. pneumophila was isolated mostly from ashiyu foot spas. Ashiyu foot spa is a type of hot spring where people bathe their feet. Ashiyu foot spa is usually in open air and freely available. Its temperature is generally controlled around 45°C. Older people often use this facility. For these people, L. pneumophila-containing aerosols generated from environmental waters could be a source of L. pneumophila infection. To the best of our knowledge, this is the first report related to isolation of L. pneumophila from ashiyu foot spa. Previous surveys of hot springs have demonstrated that around 30% of hot springs or public bathes were L. pneumophila positive [22, 23]. In this study, L. pneumophila was isolated from three of the eight sites (38%) of ashiyu foot spa sampled. These results may suggest an equivalent risk of contracting L. pneumophila at ashiyu foot spa as compared with hot spring. However, a more extensive survey is required to obtain more accurate epidemiological relevance and to analyze the risk of L. pneumophila infection from ashiyu foot spa.
The growth of the L. pneumophila isolates in liquid medium was almost the same as reference strain Lp02, but Ymt294 showed lower growth rate (Figure 1). Since the number of Ymt294 was not increased from 24 to 48 h, the growth of Ymt294 seemed to be saturated at one-tenth of final concentration of other strains. Intracellular growth of these isolates was different in HeLa and THP-1 cells. In HeLa cells, growth of isolates was significantly higher than Lp02 at 24 h after infection. However, the numbers of intracellular bacteria were decreased at 48 h after infection (Figure 2(b)). Some isolates such as Ymt294, Twr292, and Ymg289 showed strong cytotoxicity in HeLa cells, and cells were detached from culture plate at 48 h (Figures 2(c) and 4). This detachment may be a dominant factor of decrease in intracellular growth of those isolates. In THP-1 cells, the numbers of intracellular bacteria were increased from 24 to 48 h, despite the high cytotoxicity of those isolates (Figures 3(a) and 3(b)). Consistent with the strong preference of L. pneumophila for macrophages, these results indicate that macrophages are more suitable for L. pneumophila growth than epithelial cells.
Since all isolates harbored genes of well-characterized virulence factors including dot, lvh, rtx, and hsp60, the relationship between virulence factors and cytotoxicity or intracellular growth was not clear. However, the existence of genes of the virulence factors may suggest that those isolates can be human pathogenic. In particular, the Twr292 isolate from ashiyu foot spa showed high intracellular growth and strong cytotoxicity in HeLa and THP-1 cells. In addition, the contamination level of Twr292 was very high (128 CFU/100 mL). According to the guidelines of Japan's Ministry of Health, Labour and Welfare, the concentration of L. pneumophila should be maintained below 10 CFU/100 mL in hot springs or public bathes. The concentration of Twr292 was more than ten times that of the defined standard.
Overall, our results strongly suggest that ashiyu foot spa is a possible source of L. pneumophila infection.
Acknowledgment
This work was supported partially by the Ministry of Education, Science, Sports and Culture, Grant-in-Aid for Scientific Research (C), 22580333.
References
- 1.Winn WC., Jr. Legionnaires disease: historical perspective. Clinical Microbiology Reviews. 1988;1(1):60–81. doi: 10.1128/cmr.1.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila . Molecular Microbiology. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 3.Cianciotto NP. Pathogenicity of Legionella pneumophila . International Journal of Medical Microbiology. 2001;291(5):331–343. doi: 10.1078/1438-4221-00139. [DOI] [PubMed] [Google Scholar]
- 4.DebRoy S, Aragon V, Kurtz S, Cianciotto NP. Legionella pneumophila Mip, a surface-exposed peptidylproline cis-trans-isomerase, promotes the presence of phospholipase C-like activity in culture supernatants. Infection and Immunity. 2006;74(9):5152–5160. doi: 10.1128/IAI.00484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isberg RR, Rankin S, Roy CR, Swanson MS, Berger KH. Legionella pneumophila: factors involved in the route and response to an intracellular niche. Infectious Agents and Disease. 1993;2(4):220–223. [PubMed] [Google Scholar]
- 6.Marra A, Blander SJ, Horwitz MA, Shuman HA. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(20):9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Söderberg MA, Rossier O, Cianciotto NP. The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures. Journal of Bacteriology. 2004;186(12):3712–3720. doi: 10.1128/JB.186.12.3712-3720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diederen BMW. Legionella spp. and Legionnaires’ disease. Journal of Infection. 2008;56(1):1–12. doi: 10.1016/j.jinf.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Kuroki T, Ishihara T, Ito K, Kura F. Bathwater-associated cases of legionellosis in Japan, with a special focus on Legionella concentrations in water. Japanese Journal of Infectious Diseases. 2009;62(3):201–205. [PubMed] [Google Scholar]
- 10.Nakamura H, Yagyu H, Kishi K, et al. A large outbreak of Legionnaires’ disease due to an inadequate circulating and filtration system for bath water— epidemiologic manifestations. Internal Medicine. 2003;42(9):806–811. doi: 10.2169/internalmedicine.42.806. [DOI] [PubMed] [Google Scholar]
- 11.Okada M, Kawano K, Kura F, et al. The largest outbreak of legionellosis in Japan associated with spa baths: epidemic curve and environmental investigation. The Journal of the Japanese Association for Infectious Diseases. 2005;79(6):365–374. doi: 10.11150/kansenshogakuzasshi1970.79.365. [DOI] [PubMed] [Google Scholar]
- 12.Dennis P. Isolation of Legionella from environmental specimens. In: Harrison TG, Taylor AG, editors. A Laboratory Manual for Legionella. Hoboken, NJ, USA: John Wiley & Sons; 1988. pp. 31–44. [Google Scholar]
- 13.Maiwald M, Helbig JH, Lück PC. Laboratory methods for the diagnosis of Legionella infections. Journal of Microbiological Methods. 1998;33(1):59–79. [Google Scholar]
- 14.Ratcliff RM, Slavin MA, Sangster N, Doyle RM, Seymour JF, Lanser JA. Legionella pneumophila mip gene sequencing to investigate a cluster of pneumonia cases. Pathology. 2003;35(1):65–69. [PubMed] [Google Scholar]
- 15.Huang B, Yuan Z, Heron BA, et al. Distribution of 19 major virulence genes in Legionella pneumophila serogroup 1 isolates from patients and water in Queensland, Australia. Journal of Medical Microbiology. 2006;55(8):993–997. doi: 10.1099/jmm.0.46310-0. [DOI] [PubMed] [Google Scholar]
- 16.Jaulhac B, Nowicki M, Bornstein N, et al. Detection of Legionella spp. in bronchoalveolar lavage fluids by DNA amplification. Journal of Clinical Microbiology. 1992;30(4):920–924. doi: 10.1128/jcm.30.4.920-924.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturgill-Koszycki S, Swanson MS. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. Journal of Experimental Medicine. 2000;192(9):1261–1272. doi: 10.1084/jem.192.9.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal G, Russo JJ, Shuman HA. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila . Molecular Microbiology. 1999;34(4):799–809. doi: 10.1046/j.1365-2958.1999.01642.x. [DOI] [PubMed] [Google Scholar]
- 19.Cirillo SLG, Bermudez LE, El-Etr SH, Duhamel GE, Cirillo JD. Legionella pneumophila entry gene rtxA is involved in virulence. Infection and Immunity. 2001;69(1):508–517. doi: 10.1128/IAI.69.1.508-517.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez RC, Logan SM, Lee SHS, Hoffman PS. Elevated levels of Legionella pneumophila stress protein Hsp60 early in infection of human monocytes and L929 cells correlate with virulence. Infection and Immunity. 1996;64(6):1968–1976. doi: 10.1128/iai.64.6.1968-1976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garduño RA, Garduño E, Hoffman PS. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infection and Immunity. 1998;66(10):4602–4610. doi: 10.1128/iai.66.10.4602-4610.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuhata K, Hara M, Yoshida S-I, Fukuyama M. Distribution of Legionella spp. in hot spring baths in Japan. The Journal of the Japanese Association for Infectious Diseases. 2004;78(8):710–716. doi: 10.11150/kansenshogakuzasshi1970.78.710. [DOI] [PubMed] [Google Scholar]
- 23.Karasudani T, Kuroki T, Otani K, et al. Legionella contamination risk factors in non-circulating hot spring water. The Journal of the Japanese Association for Infectious Diseases. 2009;83(1):36–44. doi: 10.11150/kansenshogakuzasshi.83.36. [DOI] [PubMed] [Google Scholar]


