Fig. 1. Outline of the 4-plex iTRAQ based quantitative proteomic strategy.
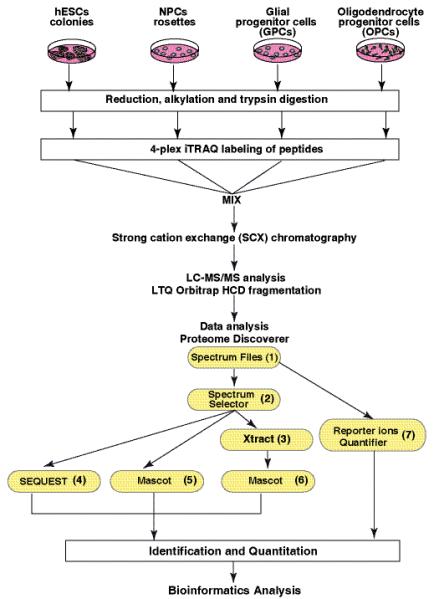
A. iTRAQ labeling was carried out using lysates from embryonic stem cells (ESC), neural progenitor cells (NPC), glial progenitor cells (GPC) and oligodendrocyte progenitor cells (OPC). Samples were digested using trypsin and labeled using iTRAQ reagents 114, 115, 116 and 117 with peptides from ESC, NPC, GPC and OPC respectively. After labeling, peptides from all four samples were combined and fractionated by SCX chromatography. Each fraction was then analyzed by LC-MS/MS on a LTQ Orbitrap mass spectrometers. The data was analyzed using Proteome Discoverer software. The workflow nodes (1 to 6) included spectrum files (raw mass spectrometry data selector), Spectrum selector, Sequest and Mascot search engines and reporter ions quantifier.
