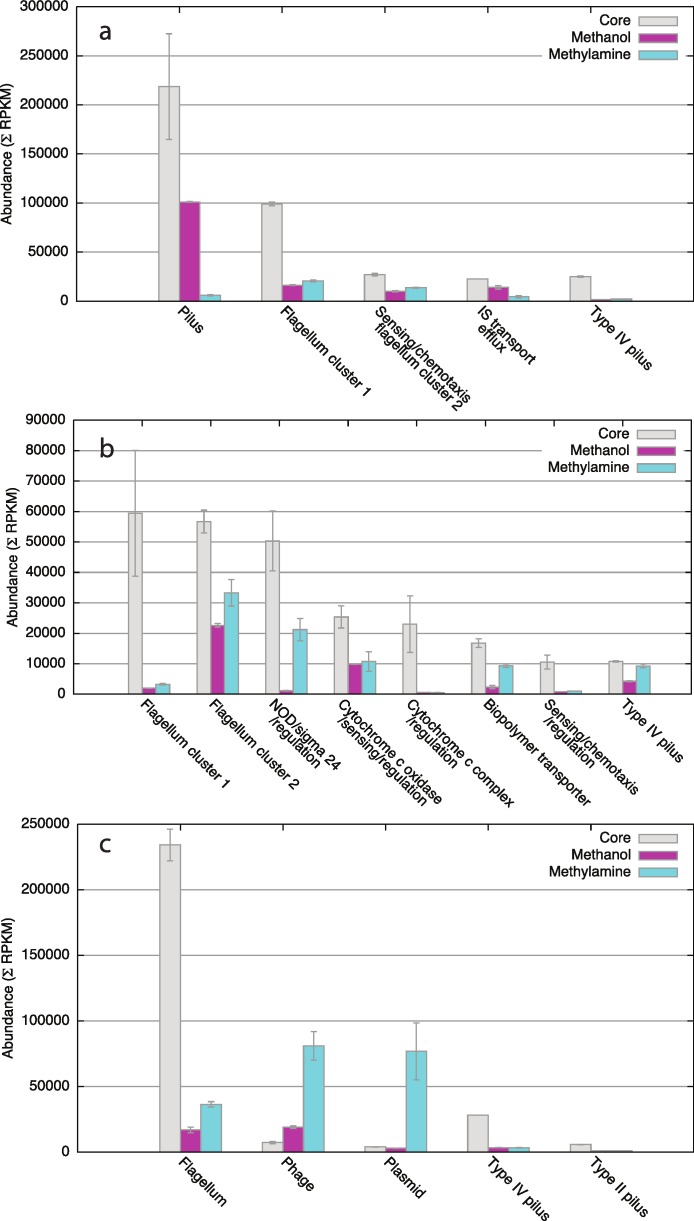
Figure 5. Combined relative abundances of transcripts for genes most highly and most differentially expressed in each of the three organisms.
(A) M. mobilis JLW8 unique pilus genes, mmol_1380-1409; flagellum cluster 1, mmol_0919-0953; sensing/chemotaxis/flagellum cluster 2, mmol_1248-1273; IS transport/efflux, mmol_1451-1493; Type IV pilus mmol_2269-2274. (B) M. versatilis 301 flagellum cluster 1, m301_1679-1716; flagellum cluster 2, m301_0961-1015; NOD/sigma 24/regulation, m301_0769-0776; cytochrome c oxidase/sensing/regulation, m301_0844-0858; cytochrome c complex/regulation, m301_0064-0069; biopolymer transporter, m301_2524-2533; sensing/chemotaxis/regulation, m301_0896-0902; Type IV pilus, m301_2616-2621. (C) M. glucosotrophus SIP3-4 flagellum, msip34_0747-0820; phage, msip34_2008-2089; plasmid, msip34_2831-2843; Type IV pilus, msip34_2593-2598; Type II pilus, msip34_2502-2507.