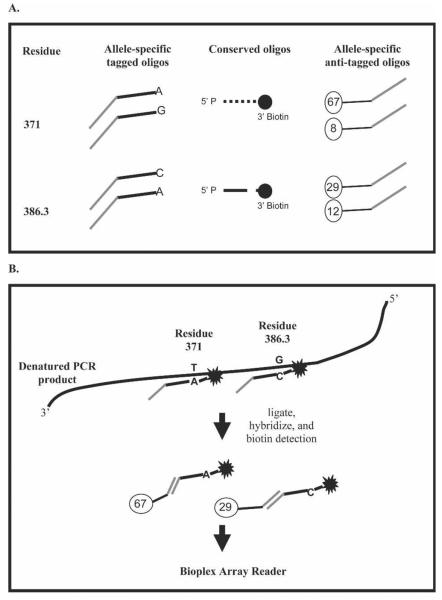
Figure 1.
Post-PCR OLA for SNP detection. P. vivax Duffy binding protein amino acid residues 371 and 386.3 are shown as examples of how SNPs can be multiplexed in the same reaction. See Table 1 for the oligonucleotide sequences. (A) Oligonucleotides necessary to type the two SNPs. Two allele-specific oligonucleotides were designed for each SNP containing a 25-nucleotide TAG at the 5′ end to allow subsequent hybridization with anti-TAG probe coupled to a unique fluorescent microsphere. Common oligonucleotides adjacent to each SNP were designed with a 5′ phosphate group and a 3′ biotin. (B) Allele-specific oligonucleotide that hybridized to the target amplicon was covalently ligated to the common oligonucleotide. Allele-specific ligation products were hybridized with the anti-TAG fluorescent microspheres, and streptavidin-R-phycoerythrin was also added to detect the biotin labeled common oligonucleotides. Detection of allele-specific bead-labeled anti-TAG hybrid complexes was performed using a Bio-Plex array reader.