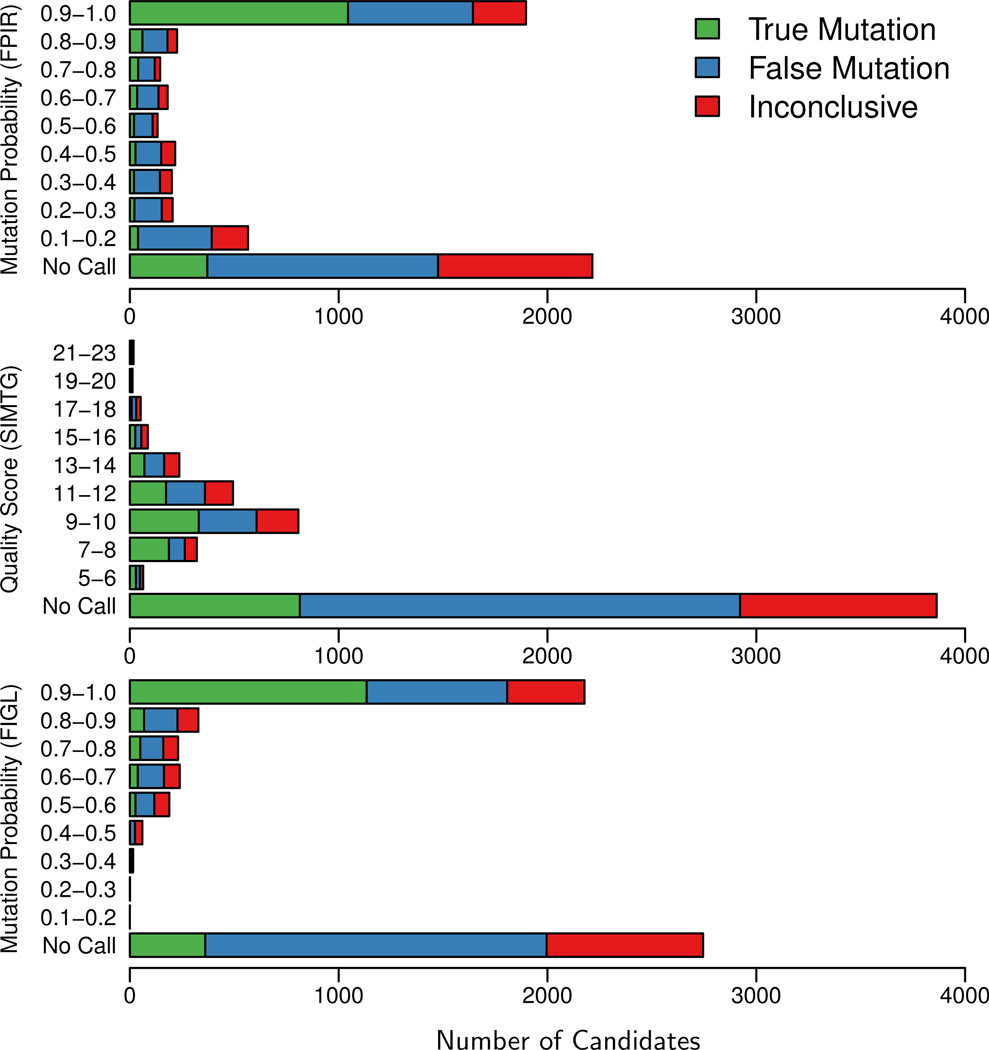
Figure 7.
Validation results stratified by de novo calling algorithm. Three methods were used to produce candidate sites of de novo mutation in two families (Conrad et al., 2011, Durbin et al., 2010). These candidates were then experimentally validated and classified as either “true mutations” (germline, somatic, or cell-line), “false mutations” (inherited genetic variation or no variation at all), or “inconclusive”. Sites not reported by a method but reported by another method appear in the No Call bar. FPIR corresponds to the method described in this paper. Modified from Conrad et al. (2011).