Figure 2. Identification of Small GTPases that Directly Activate PI3K.
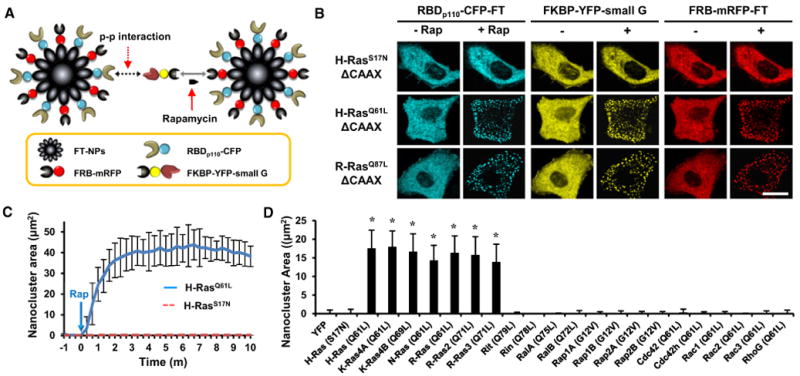
(A) Schematic diagram of the InCell SMART-i technique used to observe protein-protein interactions using ferritin-derived nanoparticles.
(B) Constitutively active H- and R-Ras bound to RBDp110. Scale bar, 20 μm.
(C) Kinetic analysis of the nanoclusters of constitutively active H-Ras (solid line) and dominant-negative H-Ras (dotted line). Data represent the mean ± SD (n> 10).
(D) After rapamycin treatment for 1 hr, the nanoclusters of the 21 constitutively active small GTPases were measured. YFP and dominant-negative H-Ras were used as negative controls. Data represent the mean ± SD (n > 20; * indicates p < 0.001 compared with control [YFP]).
