Table 1. Crystallographic data-collection and refinement statistics for Na-GST-3.
Values in parentheses are for the highest resolution shell.
| Space group | P43212 |
| Unit-cell parameters (Å, °) | a = b = 67.12, c = 134.95, α = β = γ = 90 |
| Resolution (Å) | 32.5–2.07 (2.24–2.07) |
| R merge † (%) | 10.3 (45.2) |
| Completeness (%) | 99.96 (100) |
| Multiplicity | 13.0 (12.9) |
| 〈I/σ(I)〉 | 5.7 (1.7) |
| Refinement | |
| R factor‡ (%) | 17.2 (18.3) |
| R free § (%) | 21.5 (26.4) |
| Correlation coefficients | |
| F o − F c | 0.951 |
| F o − F c, free | 0.930 |
| Components of the model | |
| Amino-acid residues | 205 |
| Waters | 72 |
| GSH | 1 |
| Glycerols | 2 |
| Sulfate | 1 |
| Mean B factor (Å2) | 24.9 |
| R.m.s. deviation from ideal | |
| Bond lengths (Å) | 0.019 |
| Bond angles (°) | 1.87 |
| Chirality (Å3) | 0.14 |
| Ramanchandran plot, No. of residues in | |
| Favored regions | 197 |
| Allowed regions | 2 |
| Outlier regions | 1 |
R
merge = 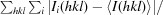
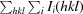 , where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
, where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
R factor = 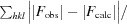
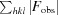 , where F
obs are observed and F
calc are calculated structure factors.
, where F
obs are observed and F
calc are calculated structure factors.
The R free set consists of a randomly chosen 5% of reflections.
