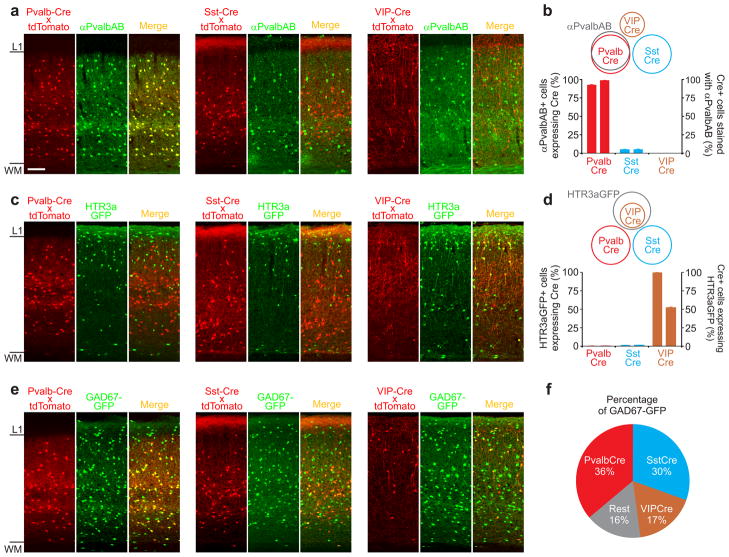
Figure 1. Three non-overlapping Cre-driver lines.
a) Confocal double fluorescence images of coronal sections through visual cortex of three Cre-driver lines (Pvalb-Cre (left); Sst-Cre (center); VIP-Cre (right)). Cre expression pattern (labeled in red, revealed by crossing the Cre-driver lines with the ROSA-tdTomato reporter line; left sub-panels) counterstained with anti Pvalb antibody (labeled in green; center sub-panels) and overlay (right sub-panels). Note labeling of Pvalb-Cre cells but not Sst-Cre or VIP-Cre cells with anti Pvalb antibodies (yellow cells in right sub-panel). Scale bar: 100μm; same for all panels in figure 1.
b) Schematic of overlap of Cre lines with respect to Pvalb antibody labeling (top) and quantification of overlap (bottom). The left and right ordinates refer to the left and right data-columns of the same cre-line, respectively. Error bars represent s.e.m. (Pvalb: n=1548 cells, 4 sections, 2 mice; Sst: n=1933 cell, 6 sections, 2 mice; VIP: n=1465 cells, 6 section, 2 mice).
c) Confocal double fluorescence images of coronal sections through visual cortex of the three Cre-driver lines crossed with the HTR3a-GFP line. Cre expression pattern (labeled in red, revealed by crossing the Cre-driver lines with the ROSA-tdTomato reporter line; left sub-panels); HTR3a-GFP (labeled green; center sub-panels) and overlay (right sub-panels). Note co-labeling of VIP-Cre cells but not Pvalb-Cre or Sst-Cre cells with GFP (yellow cells in right sub-panel).
d) Schematic of overlap of Cre lines with cells labeled in the HTR3a-GFP line (top) and quantification of overlap (bottom). The left and right ordinates refer to the left and right data-columns of the same cre-line, respectively. Error bars represent s.e.m. (Pvalb: n=1373 cells, 4 sections, 2 mice; Sst: n=1666 cells, 4 sections, 2 mice; VIP: n=1243 cells, 4 sections, 2 mice).
e) as in (c) but three Cre-driver lines crossed with the GAD67-GFP line.
f) Quantification of overlap of Cre-lines with cells labeled in the GAD67-GFP line. (Pvalb: overlap=36.2±0.5 s.e.m, n=1548 cells, 4 sections, 2 mice; Sst: overlap=30.4±1.5 s.e.m, n=3869, 6 sections, 2 mice; VIP: overlap=17.4±1 s.e.m, n=3033 cells, 6 section, 2 mice).