The synthesis of the immune signal salicylic acid is abolished by a mutation in a hitherto unknown transporter protein. This article describes the transporter localization at the chloroplast and its function in the export of salicylic acid from the chloroplast.
Abstract
Salicylic acid (SA) is central for the defense of plants to pathogens and abiotic stress. SA is synthesized in chloroplasts from chorismic acid by an isochorismate synthase (ICS1); SA biosynthesis is negatively regulated by autoinhibitory feedback at ICS1. Genetic studies indicated that the multidrug and toxin extrusion transporter ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5) of Arabidopsis (Arabidopsis thaliana) is necessary for SA accumulation after biotic and abiotic stress, but so far it is not understood how EDS5 controls the biosynthesis of SA. Here, we show that EDS5 colocalizes with a marker of the chloroplast envelope and that EDS5 functions as a multidrug and toxin extrusion-like transporter in the export of SA from the chloroplast to the cytoplasm in Arabidopsis, where it controls the innate immune response. The location at the chloroplast envelope supports a model of the effect of EDS5 on SA biosynthesis: in the eds5 mutant, stress-induced SA is trapped in the chloroplast and inhibits its own accumulation by autoinhibitory feedback.
Plants perceive pathogens such as viruses, bacteria, fungi, and oomycetes or abiotic stress and respond by a characteristic innate immune response that leads to the induction of local and systemic resistance. It concomitantly leads to the production of salicylic acid (SA), an innate immune signal responsible for transcriptional changes that result in resistance in the infected and neighboring cells (Vlot et al., 2009). The biological importance of SA was demonstrated using mutant or transgenic plants impaired in the accumulation of SA (Vlot et al., 2009). For example, Arabidopsis (Arabidopsis thaliana) ENHANCED DISEASE SUSCEPTIBILITY5 (eds5)/SA INDUCTION-DEFICIENT1 (sid1) and ics1/sid2 mutants impaired in pathogen- and UV-induced SA accumulation are susceptible to Pseudomonas syringae and Hyaloperonospora arabidopsidis (Nawrath et al., 2002); plants degrading SA upon expression of the bacterial enzyme naphthalene hydroxylase G fail to accumulate SA and SA-dependent defense responses (Gaffney et al., 1993; Vernooij et al., 1994). A mutation in the ISOCHORISMATE SYNTHASE1 (ICS1) gene (Wildermuth et al., 2001) unveiled the essential role of isochorismate in the synthesis of SA, an observation confirmed in many plants (Verberne et al., 2007; Catinot et al., 2008). SA biosynthesis is localized in the chloroplast (Strawn et al., 2007; Fragnière et al., 2011). This raises the question of how SA is exported to the cytoplasm, from where it transits to its binding site in the nucleus (Fu et al., 2012). The eds5/sid1 mutation was mapped to EDS5, a gene encoding a member of the multidrug and toxin extrusion (MATE) transporter family (Nawrath et al., 2002). MATE transporters are found in prokaryotes and eukaryotes and represent conserved protein families (Omote et al., 2006; Kuroda and Tsuchiya, 2009). The kingdom of plants has the largest number of MATE genes; for example, there are 58 MATE genes in Arabidopsis. They are often H+/cation antiporters associated with the extrusion of secondary (Debeaujon et al., 2001; Marinova et al., 2007) or toxic (Morita et al., 2009; Shoji et al., 2009) metabolites. In some cases, anions such as citrate can also be transported (Magalhaes et al., 2007; Maron et al., 2010; Yokosho et al., 2009).
But how a putative transport protein could possibly regulate SA biosynthesis has remained a riddle. Here, we experimentally test the hypothesis that EDS5 is localized at the chloroplast envelope membrane and catalyzes the export of SA from the chloroplast to the cytoplasm.
RESULTS AND DISCUSSION
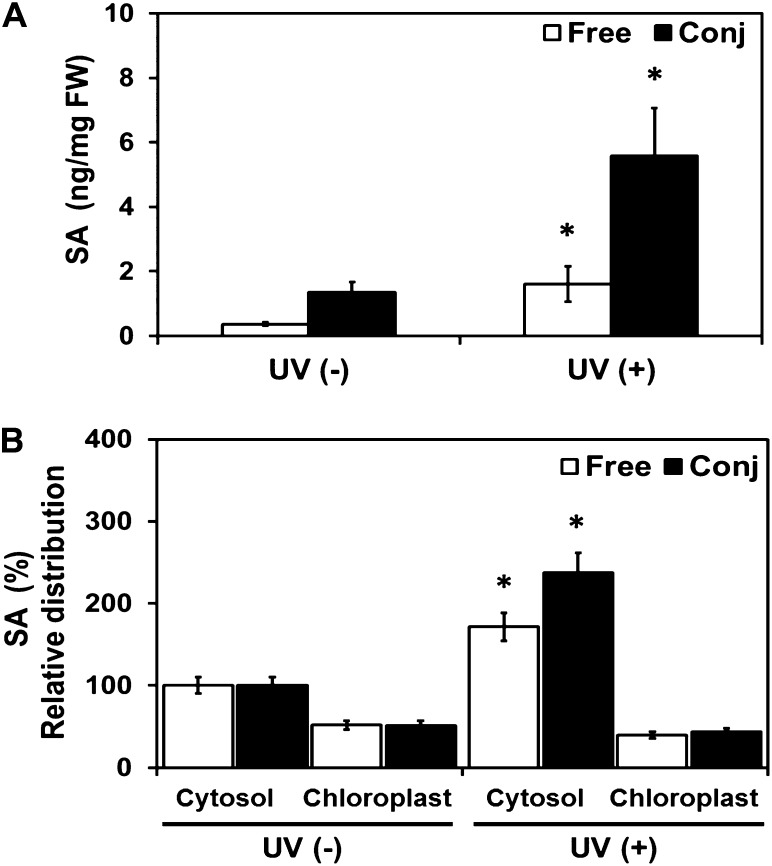
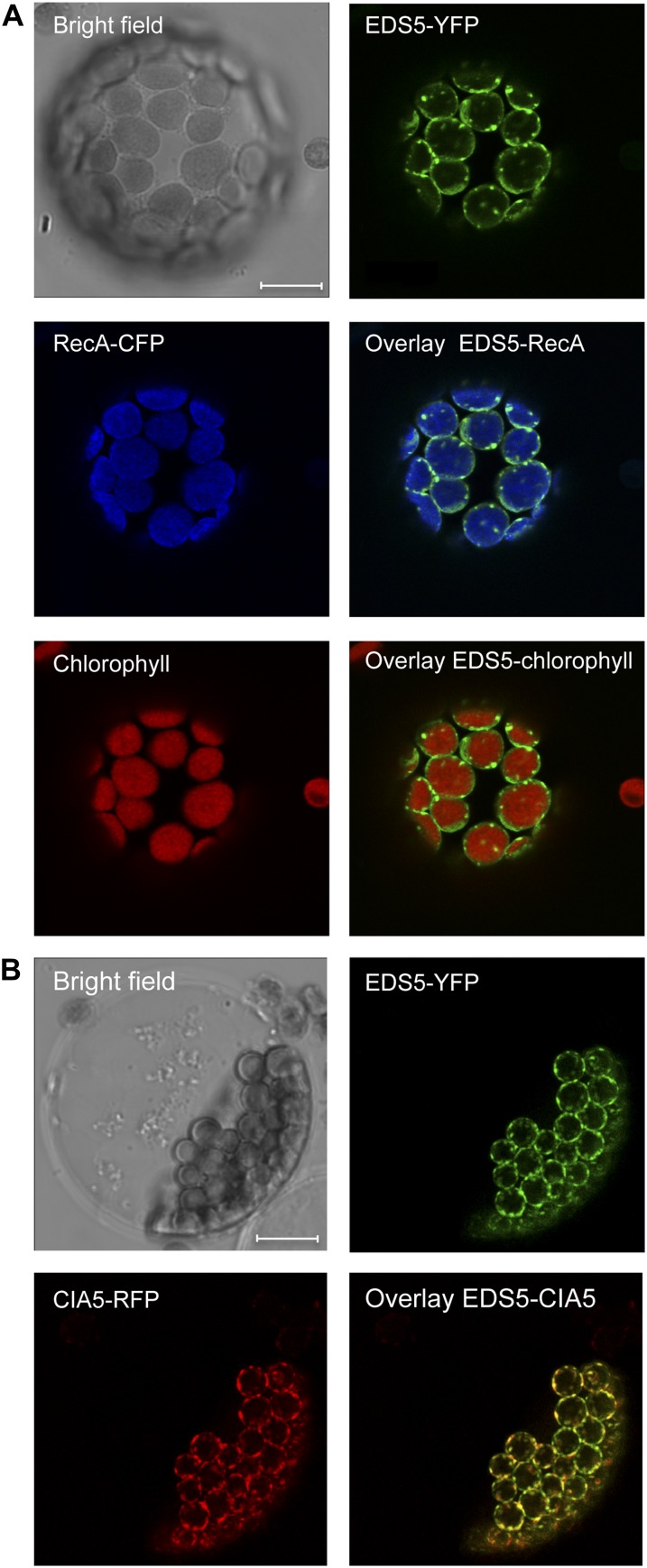
Free and conjugated SA accumulate in the leaves of Arabidopsis after stimulation with UV light (Fig. 1A), as described previously (Nawrath et al., 2002; Fragniére et al., 2011). Surprisingly the increase in free and conjugated SA is not measurable in the chloroplast fraction (Fig. 1B). This might be explained by rapid export of SA from the chloroplast to the cytoplasm. Thus, we predict that (1) EDS5 is located at the chloroplast envelope; (2) mutants with impaired EDS5 function are unable to export SA; and therefore (3) in these mutants, SA is trapped in the chloroplast and inhibits its synthesis by negative feedback. To test this hypothesis, EDS5 was localized by the stable expression of fluorescent EDS5-yellow fluorescent protein (YFP) under the control of the constitutive cauliflower mosaic virus 35S (35S) promoter (35S::EDS5-YFP) in transgenic Arabidopsis plants that were cotransformed with the chloroplast marker 35S::RecA-CFP (see “Materials and Methods”). Confocal laser scanning microscopy (CLSM) of mesophyll protoplasts resulted in RecA-cyan fluorescent protein (CFP) fluorescence overlapping with the autofluorescence of chlorophyll in plastids (Fig. 2A). In contrast, the fluorescence of EDS5-YFP localized to the plastid periphery while chlorophyll autofluorescence and RecA-CFP fluorescence appeared at the plastid center. This suggested that EDS5-YFP is localized at the plastid envelope (Fig. 2A). The envelope localization of EDS5 was confirmed by the transformation of mesophyll protoplasts of 35S::EDS5-YFP plants with the chloroplast envelope marker construct prCIA5-TM2-RFP (Teng et al., 2006), allowing for colocalization. The fluorescence of prCIA5-TM2-red fluorescent protein (RFP) overlapped perfectly with that of EDS5-YFP, indicating a localization of the latter to the chloroplast envelope (Fig. 2B).
Figure 1.
Distribution of free and conjugated SA in leaves of Arabidopsis after stimulation of SA biosynthesis by UV exposure. Three-week-old Arabidopsis plants were exposed to UV, and SA was subsequently determined. A, Free and conjugated SA content in whole leaves. B, SA relative distribution to non-UV-induced plants in the cytosol and chloroplasts. Representative results of three independent experiments each with three replicates are shown. Significant differences (Student’s t test; P < 0.05) of means ± sd from noninduced wild-type plants are indicated by asterisks. FW, Fresh weight.
Figure 2.
EDS5 localizes at the chloroplast envelope. A, CLSM images of mesophyll protoplasts from Arabidopsis expressing EDS5-YFP and the chloroplast marker RecA-CFP. B, Mesophyll protoplasts from Arabidopsis expressing EDS5-YFP transfected with the inner envelope marker prCIA5-TM2-RFP (CIA5-RFP; Cerutti et al., 1992). The protoplast isolation and transfection were carried out as described (Yoo et al., 2007). Bars = 10 μm.
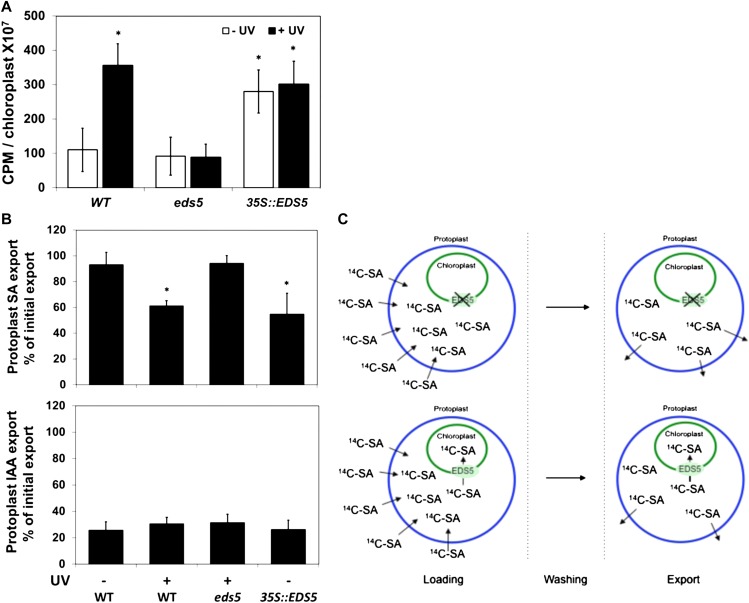
The functionality of EDS5 as an SA transporter was tested biochemically in transport assays and using a genetic approach. First, we loaded isolated chloroplasts with radiolabeled SA followed by immediate quantification (Fig. 3A). As expected, an increase in [14C]SA uptake could only be observed in plants expressing EDS after UV exposure or in transgenic plants overexpressing EDS5 (35S::EDS5). No accumulation of [14C]SA was observed in chloroplast from eds5 mutants (Fig. 3A). Second, to further support this finding, we quantified the simultaneous export of [14C]SA and [3H]3-indoleacetic acid (IAA), the latter used here as an unspecific control, from loaded entire mesophyll protoplasts. In agreement with the results obtained so far, only UV-treated or 35S::EDS5 protoplasts displayed reduced [14C]SA export over the plasma membrane, most probably due to enhanced chloroplast trapping. Interestingly, no significant difference was found for the [3H]IAA control substrate, indicating a high degree of transport specificity (Fig. 3B).
Figure 3.
EDS5 catalyzes the specific transport of SA. A, UV induction or constitutive overexpression of EDS5 enhances SA uptake into isolated chloroplasts. Chloroplasts were incubated with labeled SA ([14C]SA), and SA uptake was quantified by scintillation counting. Enhanced SA uptake caused by UV induction or constitutive overexpression of EDS5 (35S::EDS5) is absent in eds5, demonstrating the identity of EDS5 as an SA transporter. Significant differences (Student’s t test; P < 0.05) of means ± sd (n = 4) from non-UV-induced wild-type (WT) plants are indicated by asterisks. B, Export of SA but not IAA from protoplasts is reduced by UV induction or constitutive overexpression of EDS5. Mesophyll protoplasts were simultaneously loaded with [14C]SA and the auxin IAA ([3H]IAA) as an unspecific control. Reduced SA export over the protoplast plasma membrane caused by UV induction or constitutive overexpression of EDS5 (35S::EDS5) is most probably due to chloroplast trapping of SA provided by EDS5; its absence in eds5 demonstrates indirectly the identity of EDS5 as an SA transporter. EDS5 is specific for SA, as chloroplast trapping by EDS5 was not observed with IAA, used as an unspecific control. Significant differences (ANOVA using the Tukey test for multiple comparisons; P < 0.05) of means ± se (n = 4) from noninduced wild-type plants are indicated by asterisks. Note that the chloroplast uptake directionalities reported here for EDS5 in A and B underline the expected bidirectionality of EDS5 transport activity and are most likely caused by the nonphysiological high concentration of labeled SA during the transport assays. C, Schematic representation of the protoplast export experiment. Protoplasts of Arabidopsis leaves prepared from wild-type, UV-treated wild-type, 35S::EDS5, and eds5 plants were incubated with [14C]SA (left). Protoplasts were then washed (middle) and incubated in fresh medium, allowing the quantification of export (right). When EDS5 is present (after UV treatment or in 35S::EDS5 plants; bottom panel), SA is transported into the chloroplast and will contribute to a lesser total export over the plasma membrane, as observed in B.
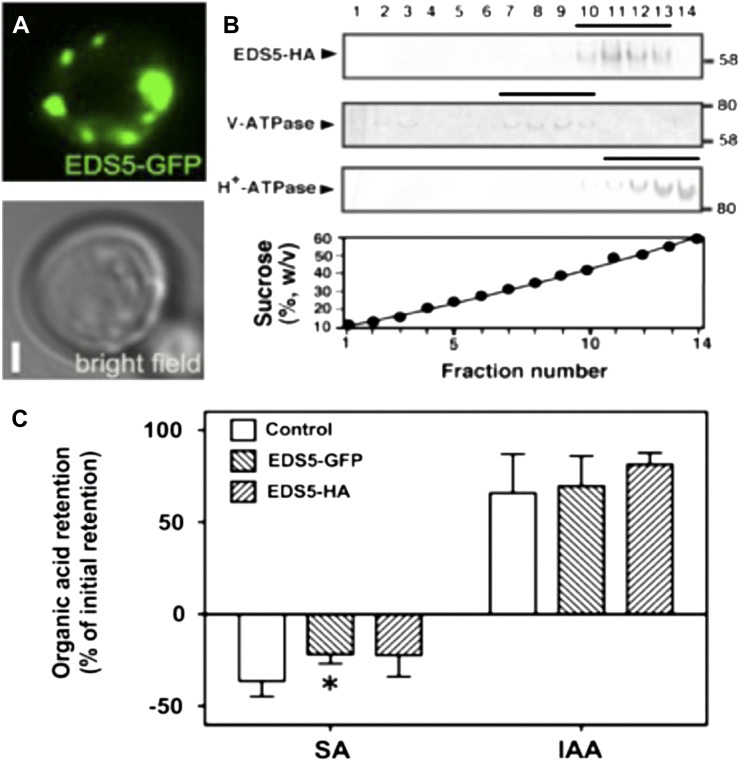
Further evidence for a direct transport activity was obtained by expressing EDS5 in bakers’ yeast (Saccharomyces cerevisiae strain JK93da; Geisler et al., 2005). Like other transporters (Bailly et al., 2008), EDS5-GFP was localized to punctate, raft-like structures surrounding the yeast plasma membrane (Fig. 4A). Localization at the plasma membrane was also confirmed by membrane fractionation. A wide overlap with the plasma membrane marker H+-ATPase was observed in membrane fractions separated on Suc density gradients followed by western-blot analysis (Fig. 4B). A plasma membrane location for EDS5 made it possible to test the transport of SA in whole yeast cells (Geisler et al., 2005). However, in analogy to chloroplast experiments, export experiments were hindered by the rapid efflux of loaded radioactive SA from yeast cells. Therefore, SA loading assays were used (Kamimoto et al., 2012). Vector control yeast cells (control in Fig. 4C) showed a negative net retention of radiolabeled SA, indicating a strong endogenous efflux activity on the yeast plasma membrane (Fig. 4C). However, both hemagglutinin (HA)- and GFP-tagged versions of EDS5 exported less SA than the vector control, although only EDS5-GFP transport was significantly different from the vector control. This finding indicates that EDS5 functions as an SA transporter at the yeast plasma membrane but in an uptake direction. As shown for protoplast assays (Fig. 3), the usage of IAA as an unspecific control in the same transport assay demonstrated EDS5 specificity for SA in yeast. In contrast to SA, IAA was loaded into yeast cells, arguing for the absence of an efficient IAA export system at the yeast plasma membrane (Geisler et al., 2005) and/or a different membrane permeability or a compartmentation of IAA.
Figure 4.
EDS5 catalyzes the specific transport of SA in yeast. A, EDS5-GFP is localized on small, punctate vesicles surrounding the plasma membrane of Saccharomyces cerevisiae. B, EDS5-HA comigrates with the plasma membrane marker H+-ATPase in continuous Suc gradients judged by western detection. Assay and marker enzymes are described by Kamimoto et al. (2012). C, EDS5 yeast show reduced export of SA. Whole yeast were loaded simultaneously with labeled SA ([14C]SA) and IAA ([3H]IAA), and net retention was quantified as described by Kamimoto et al. (2012). Altered SA but not IAA retention in EDS5-HA and EDS5-GFP yeast compared with the vector control (Control) demonstrate the identity of EDS5 as a specific SA transporter. A significant difference (Student’s t test; P < 0.05) of means ± se (n = 4) from vector control yeast is indicated by the asterisk. Note that negative retention (i.e. export) for SA not found for IAA argues for a strong background SA efflux activity by an endogenous transport system on the yeast plasma membrane (for details, see text).
In order to characterize EDS5 as a MATE-like transporter that is known to be secondarily energized, mainly by proton gradients, we repeated SA loading assays in the absence and presence of the ionophores nigericin and carbonyl cyanide m-chlorophenyl hydrazone (CCCP), known to destroy proton gradients but with different preferences for other monovalent cations. Both nigericin and CCCP strongly revert negative net SA retention (export) in vector control and EDS5-GFP yeast into a net loading, supporting that SA export over the yeast plasma membrane depends on an electrochemical, most likely proton, gradient (Supplemental Fig. S1). However, the strong yeast-endogenous SA efflux activity hindered demonstration of a secondary-active, MATE-typic energization of EDS5 in yeast using ionophores.
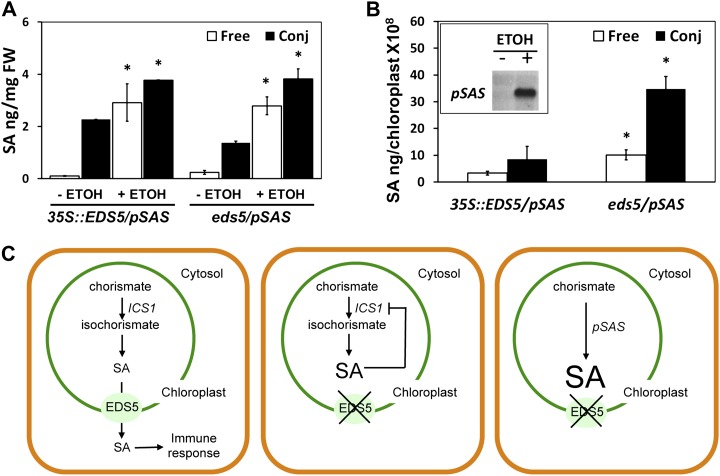
In summary, chloroplast, protoplast, and yeast experiments demonstrate that EDS5 functions as a facilitator of SA transport. Demonstration of EDS5 functionality in the extrusion of SA would require the quantification of labeled SA exported from chloroplasts. However, it was not possible to measure the export of SA from loaded chloroplasts because the labeled SA was very rapidly released by the plastids during the washing procedure. Therefore, we searched for an alternative strategy that allowed us to increase SA levels in the chloroplasts and bypass the negative feedback regulation of SA accumulation. To that end, a genetic strategy was designed whereby a chloroplast-targeted, ethanol-inducible gene encoding a chloroplast-targeted bifunctional chimeric protein referred to as SA synthase (pSAS) was engineered into eds5 mutants and EDS5-overexpressing plants (Salter et al., 1998). pSAS consists of isochorismate synthase fused to isochorismate pyruvate lyase from Pseudomonas aeruginosa (Mauch et al., 2001). Plants harboring pSAS were previously shown to produce high levels of SA in the absence of any stimulus (Mauch et al., 2001). One day after induction by ethanol, SA accumulated to similar levels in leaves of both eds5/pSAS mutants and 35S::EDS5/pSAS plants (Fig. 5A). However, the subcellular distribution of the newly synthesized SA differed dramatically. The chloroplasts of eds5/pSAS mutants contained increased levels of SA, whereas basal levels of SA were observed in the chloroplasts of 35S::EDS5/pSAS plants (Fig. 5B). SA cannot be exported from the chloroplasts in the absence of functional EDS5, demonstrating the function of this MATE-like transporter in this process.
Figure 5.
Free and conjugated SA accumulate in the chloroplasts of eds5 mutants overproducing SA. SA content in 35S::EDS5 plants and eds5 mutants were both transformed with ALC::pSAS. A, Free and conjugated SA content in leaves 0 and 24 h after treatment with ethanol. Significant differences (Student’s t test; P < 0.05) of means ± sd (n = 4) from non-ethanol-induced plants are indicated by asterisks. FW, Fresh weight. B, Free and conjugated SA content in isolated chloroplasts 24 h after treatment with ethanol. The inset shows the expression of ALC::pSAS in leaves of transgenic plants after ethanol treatment. Significant differences (Student’s t test; P < 0.05) of means ± sd (n = 4) from 35S::EDS5 plants are indicated by asterisks. C, Model of EDS5 action. Left, the functional EDS5 (in either 35S::EDS5 or wild-type plants induced by biotic or abiotic stress) exports SA made in the chloroplast. Middle, in eds5 mutants, SA accumulates in the chloroplast and presumably shuts down its own biosynthesis by a negative feedback that has yet to be characterized in detail. Right, in eds5 mutants induced to express pSAS, SA accumulates in the chloroplasts.
Our finding that both free and conjugated SA accumulated in chloroplasts of UV-treated leaves or in untreated eds5/pSAS mutants deserves a comment. So far, although various glucosyltransferases have been described in chloroplasts (Doi et al., 1966; Paquette et al., 2003), no SA conjugating system has been reported for this organelle. Most of the SA is presumably converted into SA O-β-glucoside by an SA glucosyltransferase likely to be located in the cytosol (for review, see Vlot et al., 2009). For the other reported forms of SA conjugates, including methyl-SA, methyl SA O-β-glucoside, and salicyloyl Glc ester, the site of conjugation is unknown. Our results clearly show the occurrence of a conjugated form of SA in chloroplasts, but further work will now be needed to determine the chemical nature of the conjugated forms as well as the conjugating enzyme.
Taken together, our results support the hypothesis that EDS5 is a MATE-like SA transporter located at the chloroplast envelope (Fig. 2), where it functions in the export of the innate immune signal SA (Fig. 5). The lack of SA synthesis in eds5 mutants might be explained by a possible autoinhibitory feedback regulation of SA biosynthesis in the chloroplast, but the details of this inhibition remain to be determined (a model is presented in Fig. 5C). This study provides a long-awaited answer to the question of the hitherto unknown function of the MATE-like transporter EDS5 in the control of SA during the innate immune responses in plants. It is important to note that plants obviously have recruited proteins, such as EDS5, for the control of innate immunity at the chloroplast that have structural homologs in prokaryotes, archea, and lower eukaryotes. The regulation of EDS5 in response to the perception of a wide number of biotic and abiotic stimuli all leading to SA accumulation now warrants further studies.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and UV-C Treatment
Arabidopsis (Arabidopsis thaliana) accession Columbia-0 seeds were grown on a pasteurized soil mix of humus and Perlite (3:1). Seeds were kept at 4°C for 2 d and then transferred to the growth chamber. Plants were grown during 3 weeks in a 12-h-light/12-h-dark cycle with 60% to 70% relative humidity and a day temperature of 20°C to 22°C and a night temperature of 16°C to 18°C. Wild-type plants were obtained from the Nottingham Arabidopsis Stock Centre. The Arabidopsis mutant eds5 used in all the experiments was eds5-3 (Nawrath et al., 2002). Plants were exposed to UV-C light (254 nm) for 20 min at a distance of 30 cm using a tl-900 8-W lamp (CAMAG; http://www.camag.com; Fragnière et al., 2011) and then placed in continuous light for 24 h. SA was determined in leaves or in the chloroplasts isolated from leaves 24 h after UV irradiation.
Chloroplast Preparation, SA Uptake Experiments, and Quantification of SA
The fractionation into chloroplast and cytosol and chloroplast isolation were carried out on leaves of 3-week-old Arabidopsis plants as described previously (Kubis et al., 2008). Chloroplasts were prepared as described previously (Kubis et al., 2008). They were then incubated in the extraction solution containing labeled [7-14C]SA, with specific activity of 47 Ci mmol−1. After 4 min at 4°C, the chloroplasts were filtered using AR200 silicone oil and subsequently placed in a vial for radioactive counting. SA extraction and measurement were performed as described previously (Fragnière et al., 2011).
Constructs
35S:EDS5:YFP
The EDS5 coding sequence was amplified with primers eds5-11 (5′-CACCATGCTAATCAAATCCCAAAGA-3′) and eds5-12 (5′-AATGGATTTAATCTTCTCCAC-3′) and cloned into the pENTR/D-TOPO vector, giving plasmid pENTR-107. This construct was recombined with Gateway vector pB7YWG2 (Plant Systems Biology, Ghent University) to give the pB109 (35S:EDS5:YFP) construct.
35S:RecA:CFP
The AtRecA sequence was amplified from genomic DNA using primers AtRecA-1 (5′-CACCATGGATTCACAGCTAGTCTTG-3′) and AtRecA-2 (5′-ATCGAATTCAGAACTGATTTT-3′). The resulting PCR fragment was cloned into the pENTR/D-TOPO vector to give the pENTR-RecA construct. This plasmid was recombined with pH7CWG2, leading to plasmid pH301 (35S:RecA:CFP). 35S:EDS5:YFP and 35S:RecA:CFP double transformants were generated by using a mixture of Agrobacterium tumefaciens strain GV3101 cells containing the plasmids pB109 and pH301. eds5 plants were transformed using the floral dip method (Clough and Bent, 1998) and analyzed for complementation (Supplemental Fig. S2).
35S::EDS5
The EDS5 coding sequence was amplified using the primers EDS5-s (5′-CGAATTCATGCTAATCAAATCCCAAAGATT-3′) and EDS5-as (5′-ACACCCGGGCTAAATGGATTTAATCTTCTCCAC-3′). This PCR fragment was cloned into the pGemT-Easy vector to give the pGemT-EDS5 construct. Then, EDS5 was cloned into the pART7 vector containing the 35S promoter and the OCS terminator, using the restriction enzyme EcoRI, to give the pART7-EDS5 construct. Subsequently, by digestion with NotI, the fragment 35S::EDS5-OCS was cloned into the binary vector pART27 and introduced into A. tumefaciens strain GV3101. eds5 plants were transformed using the floral dip method (Clough and Bent, 1998) and analyzed for complementation (Supplemental Fig. S1).
Yeast EDS5:GFP and EDS5-HA
EDS5 cDNA was recombined from pENTR-107 with yeast Gateway shuttle vectors, pGPD:GFP and pGPD:HA (Plant Systems Biology, Ghent University), to give pGPD:EDS5-GFP and pGPD:EDS5-HA (EDS5-GFP and EDS5-HA) constructs.
BinSRNA-pSAS
The alc gene expression system (Salter et al., 1998) was used to express the plastid-targeted pSAS encoding a chimeric salicylate synthase (Mauch et al., 2001) under the control of an ethanol-inducible promoter. The alc gene expression system is composed of two elements: the alcR encoding the transcription factor (ALCR) and a promoter derived from alcA (alcohol dehydrogenase I) from Aspergillus nidulans. alcR controls the activation of the alcA promoter in the presence of ethanol. To this end, a SalI fragment containing pSAS was cloned into the vector pACN digested with SalI. The resulting AlcA-driven expression cassette was cloned as a HindIII fragment into the HindIII-digested binary plasmid BinSRNACatN. The resulting BinSRNA-pSAS plasmid was transformed into A. tumefaciens strain GV3101 by the floral dip method (Clough and Bent, 1998).
Protoplast Isolation, Transfection, and Transport Measurements
Protoplast isolation and transfection were carried out as described (Geisler et al., 2005; Yoo et al., 2007). Protoplasts were simultaneously loaded with [14C]SA and the auxin, IAA ([3H]IAA), as an unspecific control. External radioactivity was removed by Percoll gradient centrifugation, and SA/IAA export was quantified by silicon oil centrifugation as described previously (Geisler et al., 2005). Relative export is calculated from exported radioactivity as follows: (radioactivity in the supernatant at time [t] = 10 min) − (radioactivity in the supernatant at t = 0) × 100%/(radioactivity in the supernatant at t = 0 min). Yeast transport assays with [14C]SA and [3H]IAA ([5-3H]IAA, with specific activity of 1 mCi mmol−1 [American Radiolabeled Chemicals]) were performed as described by Kamimoto et al. (2012); the ionophore concentrations were 1 μg mL−1 nigericin and 2 μm CCCP.
CLSM
CFP, YFP, and RFP fluorescence were detected using an excitation of 458 nm with a 468- to 510-nm band-pass filter, an excitation of 514 nm with a 524- to 550-nm band-pass filter, and an excitation of 561 nm with a 571- to 610-nm band-pass filter, respectively. Autofluorescence of chloroplasts was visualized using an excitation of 633 nm with a 680- to 730-nm band-pass filter. CLSM was performed on a Leica TCS SP5 AOBS confocal microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF416569.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of ionophores.
Supplemental Figure S2. Complementation analysis.
Acknowledgments
Linda Grainger, Laurence Charrier, and Antony Buchala are thanked for their technical assistance and Roger Schneiter for anti-V-ATPase.
Glossary
- SA
salicylic acid
- MATE
multidrug and toxin extrusion
- YFP
yellow fluorescent protein
- CLSM
confocal laser scanning microscopy
- CFP
cyan fluorescent protein
- IAA
3-indoleacetic acid
- HA
hemagglutinin
- pSAS
salicylic acid synthase
- RFP
red fluorescent protein
References
- Bailly A, Sovero V, Vincenzetti V, Santelia D, Bartnik D, Koenig BW, Mancuso S, Martinoia E, Geisler M. (2008) Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J Biol Chem 283: 21817–21826 [DOI] [PubMed] [Google Scholar]
- Catinot J, Buchala A, Abou-Mansour E, Métraux JP. (2008) Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett 582: 473–478 [DOI] [PubMed] [Google Scholar]
- Cerutti H, Osman M, Grandoni P, Jagendorf AT. (1992) A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci USA 89: 8068–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M. (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13: 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Doi K, Nikuni Z. (1966) ADP-D-glucose-α-i,4-glucan α-4-glucosyltransferase in spinach chloroplasts: partial purification and some properties of the enzyme. Biochim Biophys Acta 113: 312–320 [DOI] [PubMed] [Google Scholar]
- Fragnière C, Serrano M, Abou-Mansour E, Métraux JP, L’Haridon F. (2011) Salicylic acid and its location in response to biotic and abiotic stress. FEBS Lett 585: 1847–1852 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al. (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Kamimoto Y, Terasaka K, Hamamoto M, Takanashi K, Fukuda S, Shitan N, Sugiyama A, Suzuki H, Shibata D, Wang B, et al. (2012) Arabidopsis ABCB21 is a facultative auxin importer/exporter regulated by cytoplasmic auxin concentration. Plant Cell Physiol 53: 2090–2100 [DOI] [PubMed] [Google Scholar]
- Kubis SE, Lilley KS, Jarvis P. (2008) Isolation and preparation of chloroplasts from Arabidopsis thaliana plants. Methods Mol Biol 425: 171–186 [DOI] [PubMed] [Google Scholar]
- Kuroda T, Tsuchiya T. (2009) Multidrug efflux transporters in the MATE family. Biochim Biophys Acta 1794: 763–768 [DOI] [PubMed] [Google Scholar]
- Magalhaes JV, Liu J, Guimarães CT, Lana UGP, Alves VMC, Wang YH, Schaffert RE, Hoekenga OA, Piñeros MA, Shaff JE, et al. (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39: 1156–1161 [DOI] [PubMed] [Google Scholar]
- Marinova K, Pourcel L, Weder B, Schwarz M, Barron D, Routaboul JM, Debeaujon I, Klein M. (2007) The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19: 2023–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron LG, Piñeros MA, Guimarães CT, Magalhaes JV, Pleiman JK, Mao C, Shaff J, Belicuas SNJ, Kochian LV. (2010) Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J 61: 728–740 [DOI] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Gaille C, Kull B, Haas D, Reimmann C. (2001) Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J 25: 67–77 [DOI] [PubMed] [Google Scholar]
- Morita M, Shitan N, Sawada K, Van Montagu MCE, Inze D, Rischer H, Goossens A, Oksman-Caldentey KM, Moriyama Y, Yazaki K (2009) Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc Natl Acad Sci USA 106: 2447–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP. (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. (2006) The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci 27: 587–593 [DOI] [PubMed] [Google Scholar]
- Paquette S, Møller BL, Bak S. (2003) On the origin of family 1 plant glycosyltransferases. Phytochemistry 62: 399–413 [DOI] [PubMed] [Google Scholar]
- Salter MG, Paine JA, Riddell KV, Jepson I, Greenland AJ, Caddick MX, Tomsett AB. (1998) Characterisation of the ethanol-inducible alc gene expression system for transgenic plants. Plant J 16: 127–132 [Google Scholar]
- Simm S, Papasotiriou DG, Ibrahim M, Leisegang MS, Müller B, Schorge T, Karas M, Mirus O, Sommer MS, Schleiff E. (2013) Defining the core proteome of the chloroplast envelope membranes. Front Plant Sci 4: 11, 10.3389/fpls.2013.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Inai K, Yazaki Y, Sato Y, Takase H, Shitan N, Yazaki K, Goto Y, Toyooka K, Matsuoka K, et al. (2009) Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol 149: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn MA, Marr SK, Inoue K, Inada N, Zubieta C, Wildermuth MC. (2007) Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J Biol Chem 282: 5919–5933 [DOI] [PubMed] [Google Scholar]
- Teng YS, Su YS, Chen LJ, Lee YJ, Hwang I, Li HM. (2006) Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. Plant Cell 18: 2247–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberne MC, Sansuk K, Bol JF, Linthorst HJM, Verpoorte R. (2007) Vitamin K1 accumulation in tobacco plants overexpressing bacterial genes involved in the biosynthesis of salicylic acid. J Biotechnol 128: 72–79 [DOI] [PubMed] [Google Scholar]
- Vernooij B, Friedrich L, Morse A, Reist R, Kolditz-Jawhar R, Ward E, Uknes S, Kessmann H, Ryals J. (1994) Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6: 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Yokosho K, Yamaji N, Ueno D, Mitani N, Ma JF. (2009) OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol 149: 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]