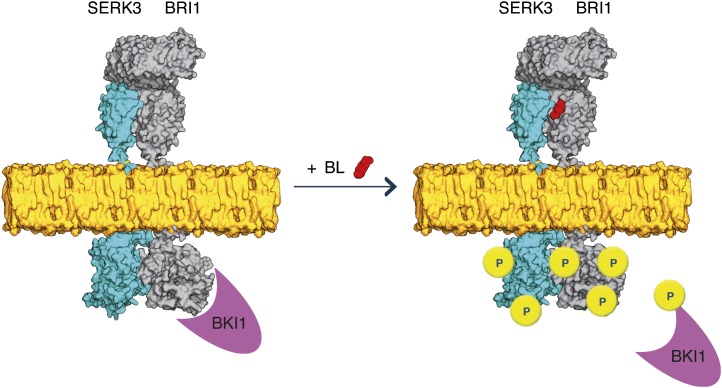
Figure 5.
Model for BR signaling with preformed BRI1-SERK3 heterooligomers as functional units. In the absence of ligand, BRI1 and SERK3 reside in the plasma membrane of Arabidopsis meristematic epidermal root cells partially as heterooligomers. The kinase activity of BRI1 is inhibited by BKI1. Upon ligand binding, the BRI1 kinase is activated. This results in transphosphorylation and release of BKI1 and transphosphorylation events within the preformed BRI1-SERK3 heterooligomers, enabling downstream signaling. Besides preformed heterooligomers, the formation of new BRI1-SERK3 complexes is possible. In this illustration, the actual protein dimensions of BRI1 and SERK3 are taken into account. The extracellular domain of SERK3 was modeled using the crystal structure of the ligand-binding domain of BRI1 (Protein Data Bank no. 3rgz) as template, and the kinase domains of SERK3 and BRI1 were modeled using the crystal structure of Pto (Protein Data Bank no. 2qkw) as template (Modeler; version 9.10). The molecular surface representation of both molecules was generated using PyMOL (version 1.4).