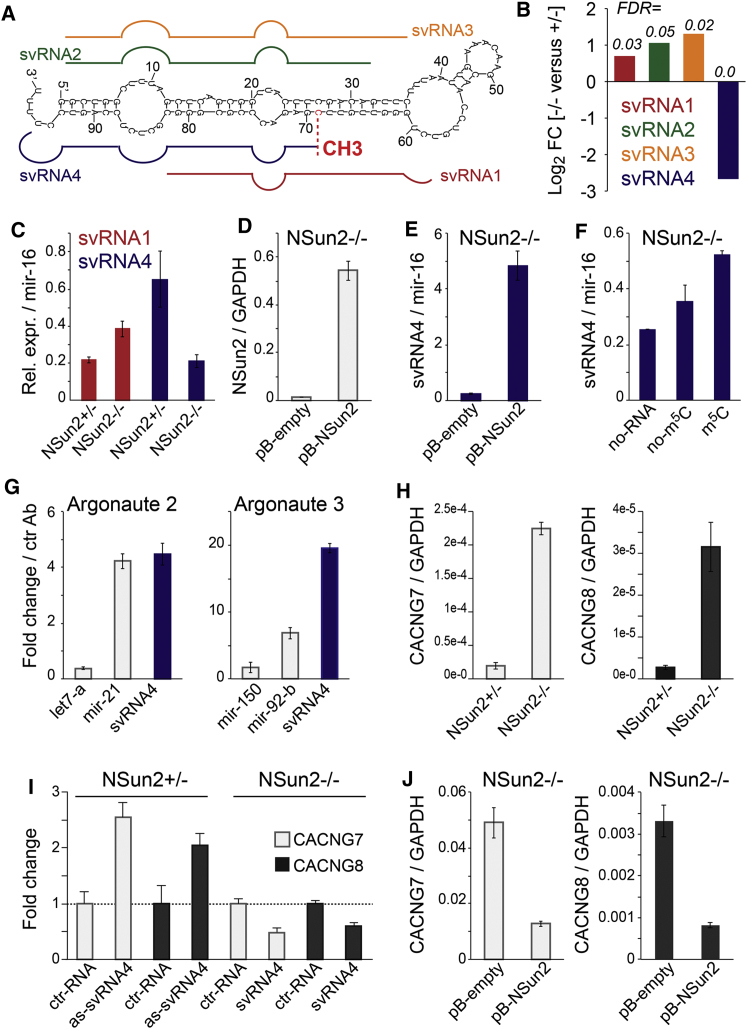
Figure 4.
Differential Processing of vtRNA1.1 into svRNAs in the Absence of m5C
(A) Schematics of secondary structure of vtRNA1.1 and small vault RNA (svRNA) found to be differentially abundant in NSUN2+/− and NSUN2−/− fibroblasts. CH3, cytosine-5 methylated site at position 69.
(B) Fold-change (log2) and false discovery rate (FDR) values for reads of svRNA1-4 in NSUN2+/− versus NSUN2−/− human fibroblasts.
(C) Detection of svRNA1 and svRNA4 in NSUN2+/− and NSUN2−/− cells using qPCR.
(D and E) RNA levels of NSun2 (D) and svRNA4 (E) in NSun2 null (−/−) fibroblasts rescued by viral infection of NSun2 (pB-NSun2) compared to the empty vector control (pB-empty).
(F) Abundance of svRNA4 in NSUN2−/− cell lysates (no-RNA) or incubated with synthetic vtRNA1.1 carrying (m5C) or lacking (no-m5C) at position 69.
(G) Detection of svRNA4 in small RNA pool copurified with Argonaute 2 (left) and Argonaute 3 (right). Let7-a and mir-150 are negative and mir-21 and mir-92-b are positive controls for Argonaute 2- and 3-bound microRNAs, respectively.
(H) qPCR showing expression of CACNG7 and CACNG8 RNA relative to GAPDH in NSUN2−/− and NSUN2+/− fibroblasts.
(I) Fold-change expression of CACNG7 and CACNG8 RNA in NSUN2−/− and NSUN2+/− fibroblasts transduced with svRNA antagomirs (as-svRNA4) or svRNA4 microRNA mimics (svRNA) versus respective control RNAs (ctr-RNA).
(J) RNA levels of CACNG7 and CACNG8 relative to GAPDH in NSun2 null (−/−) fibroblasts rescued by viral infection of NSun2 (pB-NSun2) compared to the empty vector control (pB-empty). Error estimates represent SEM (C–J).
See also Figures S5 and S6.