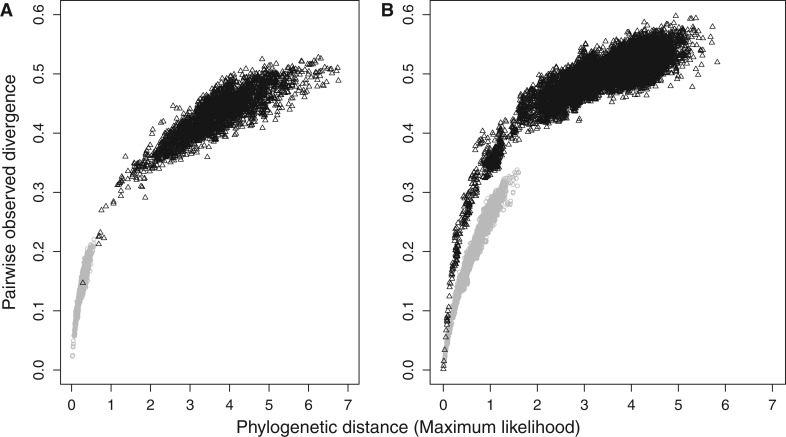
Fig. 1.—
Pairwise uncorrected divergence against phylogenetic distance estimated on the bird (A) and on the placental (B) data sets. On both panels, gray dots correspond to measures based on amino acids and black triangles to measures based on nucleotides. Phylogenetic distances are estimated by maximum likelihood using CODEML (Yang 2007; mtmam.dat substitution matrix) for the amino acid data, and BASEML (GTR+GAMMA substitution model) for nucleotides.