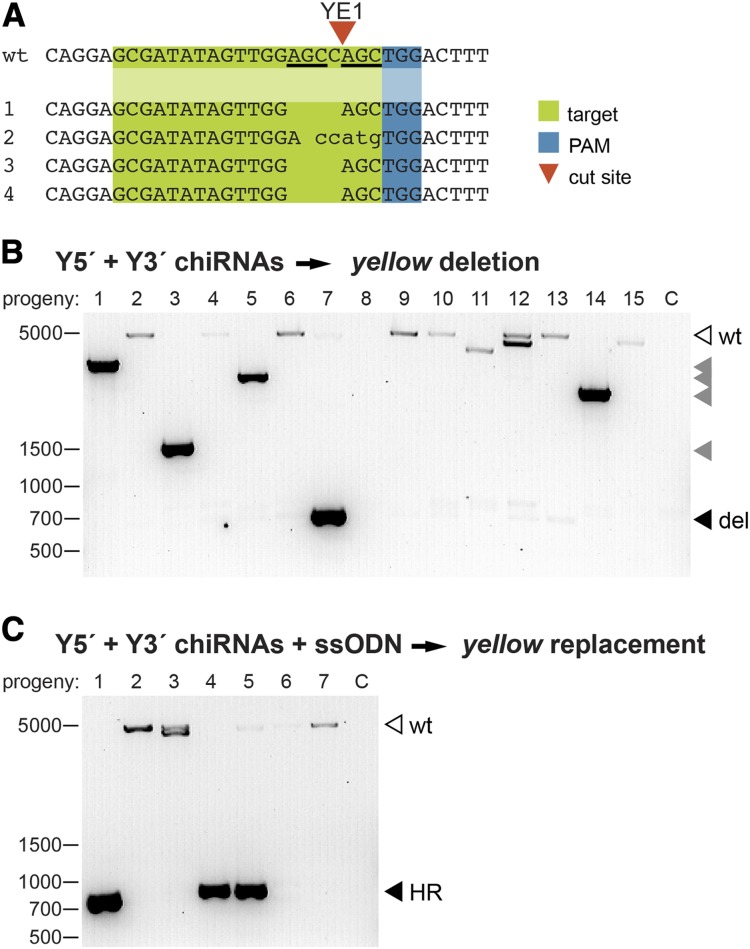
Figure 2.
CRISPR-induced modifications are efficiently transmitted through the germline. (A) Representative sequences of independent yellow mutants generated by Y1E-guided Cas9-induced DSBs. The predicted YE1 cleavage site is indicated with a red arrowhead. Short sequence repeats flanking the cleavage site (underlined in the wild-type sequence) may have facilitated the preferential generation of the same 4-bp deletion in three independent founders by MMEJ. (B) The yellow locus was amplified from yellow progeny of males and females injected with Cas9, Y5′ chiRNA, and Y3′ chiRNA using the primers indicated in Figure 1A. Fifteen independent progeny are represented (C, no template control). A 650-bp PCR product is expected (solid arrowhead) if a targeted deletion of yellow has been transmitted. PCR products corresponding to partial deletions (shaded arrowheads), and wild-type (open arrowhead) yellow are indicated. (C) The yellow locus was amplified from yellow progeny of flies injected with Cas9, Y5′ chiRNA, Y3′ chiRNA, and ssODN using the primers indicated in Figure 1A. Seven independent progeny are represented (C, no template control). A 700-bp product is expected following the targeted replacement of yellow with attP. PCR products corresponding to correct HR events are indicated (solid arrowheads) and were confirmed by Sanger sequencing.