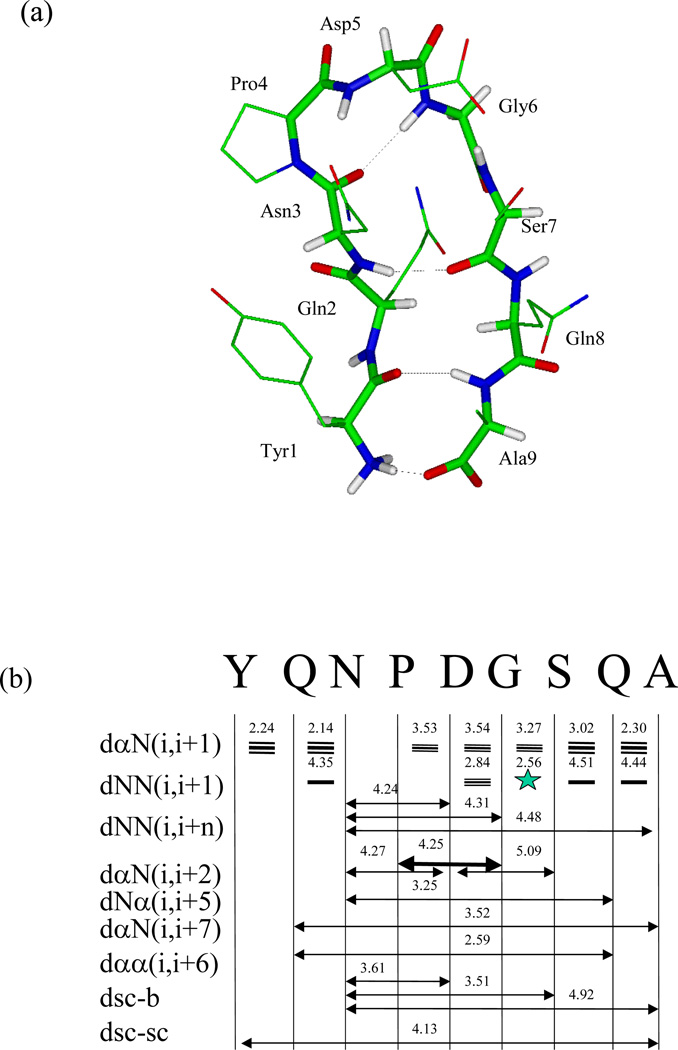
Fig. 23.
Reversible b-hairpin folding simulation with SGMD (Wu & Brooks 2004; Wu et al 2002). (a) A typical folded structure of the peptide obtained in the simulation (at 21,000 ps). For clarity, side chain hydrogens are not shown. The backbone atoms are shown as thick sticks and side chain atoms as thin sticks. Interstrand hydrogen bonds are marked by dashed lines. Atoms are colored red for oxygen, blue for nitrogen, white for hydrogen, and green for the rest. (b) NMR NOEs observed in the peptide aqueous solution2 (arrow bars between residues) and the average hydrogen pair distances (numbers in Å above NOE bars) in the β-hairpin structure obtained in our simulation. α, N, sc, and b represent the hydrogen atoms on α-carbon, amide nitrogen, side chain (β-carbon in our calculation), and backbone (amide nitrogen in our calculation). The thickness of the NOE bars represents the strength of the NOEs reported. Generally, NOEs are strong for hydrogen pair distances within 3 Å, medium between 3 and 4 Å, and week between 4 and 5 Å.