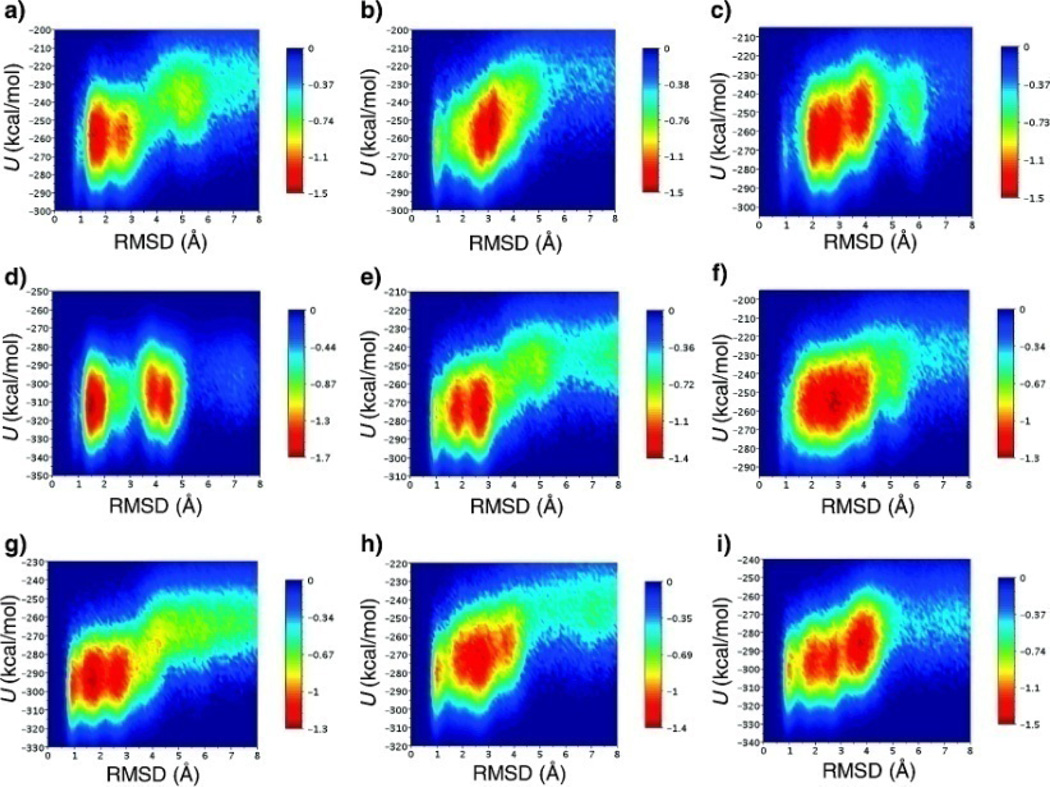
Fig 24.
Combined SGLD with replica-exchange in protein folding simulation (Lee & Olson 2010). Free-energy landscapes at respective melting temperatures (ΔGfold = 0) of individual simulations method/starting structure/simulation data〉 in the coordinates of potential energy, U, and Cα rmsd to native: (a) MD-ReX/trans/50–100 ns (T = 351.3 K), (b) MD-ReX/native/50–100 ns (T = 354.6 K), (c) MD-ReX/native/150–200 ns (T = 348.2 K), (d) LD-ReX/trans/50–100 ns (T = 290.1 K), (e) LD-ReX/native/50–100 ns (T = 335.1 K), (f) LD-ReX/native/150–200 ns (T = 353.9 K), (g) SGLD-81 ReX/trans/50–100 ns (T = 311.2 K), (h) SGLD-ReX/native/50–100 ns (T = 331.5 K), and (i) SGLDReX/native/150–200 ns (T = 306.4 K).