Figure 2. End displays stronger AP endonuclease activity in comparison to XthA.
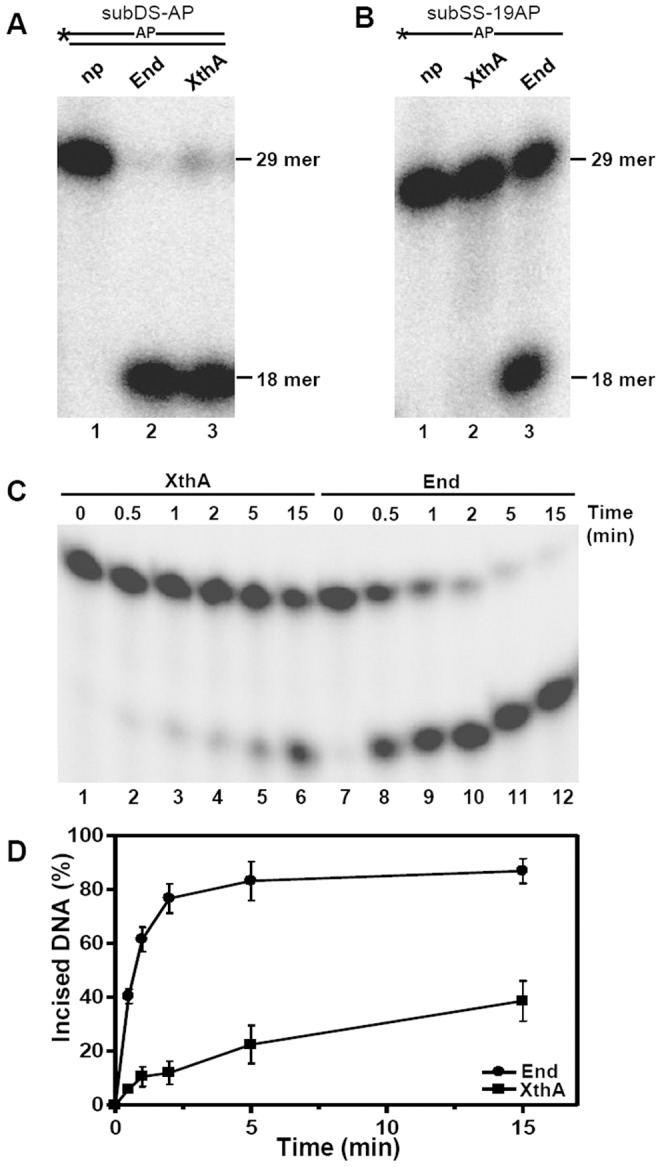
Schematic diagram of oligonucleotide substrates in which asterisk (*) and AP denote the radioactively labeled γ-32P terminus and the abasic site in the substrate, respectively. 5′ end-labeled substrate (200 fmol) was incubated with 5 fmol of either End or XthA at 37°C. (A) Lanes 1–3, subDS-AP was incubated without protein (np), with End or with XthA, respectively, for 30 min. (B) Lanes 1–3, subSS-AP was incubated without protein, with XthA or with End, respectively, for 30 min. (C) Time dependence of End and XthA on AP endonuclease activity. The reaction was carried out for different intervals of time (0–15 min). The products were analyzed by denaturing PAGE on 20% polyacrylamide gels containing 8 M Urea, and the DNA bands were visualized by autoradiography. The position of 29 mer substrate and 18 mer incision product is indicated. (D) Graphical representation of results in C. The data in the graph represent mean (±standard errors) of two independent experiments carried out in duplicates.
