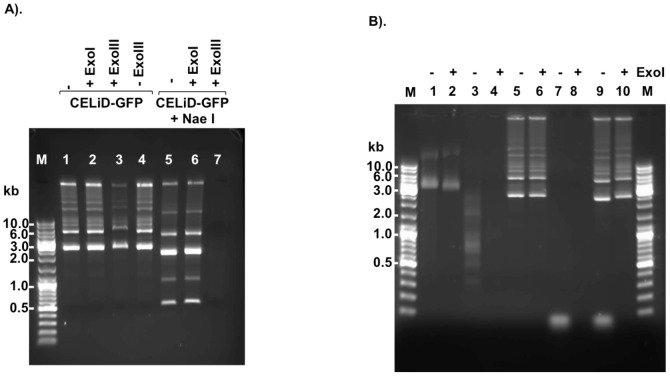
Figure 5. CELiD-GFP DNA sensitivity to exonuclease I and exonuclease III.
E. coli exonuclease III (ExoIII) removes nucleotides processively (3′ –>5′) from DNA initiating at a 3′-OH of either blunt-ended or 5′ protruding duplex DNA. E. coli exonuclease I (ExoI) degrades single-stranded DNA processively in a 3′ to 5′ direction. (A) Total CELiD DNA (1 µg) was incubated with either ExoI (20 units) or ExoIII (100 units), either without prior restriction enzyme (RE) digestion (left) or following RE digestion (right). Lane M: DNA size ladder. Lane 1: untreated CELiD DNA. Lane 2: CELiD DNA treated with ExoI. Lane 3: CELiD DNA treated with ExoIII. Lane 4: untreated CELiD DNA. Samples in lanes 5, 6, 7 were digested with NaeI prior to exonuclease treatment. Lane 5: no exonuclease control. Lane 6: ExoI treatment. Lane7: ExoIII treatment. (B) Additional substrates as controls for ExoI activity. The φX174 virus genome is a 5386 nt, single-stranded, closed circular DNA molecule. ExoI treatment is indicated above each lane by a “+” or “–” sign. Lanes 1 and 2: φX174 DNA. Lanes 3 and 4: φX174 DNA digested with HaeIII. Lanes 5 and 6: CELiD DNA. Lanes 7 and 8: single-stranded synthetic oligonucleotide (25mer). Lanes 9 and 10: Mixed CELiD DNA and 25mer.