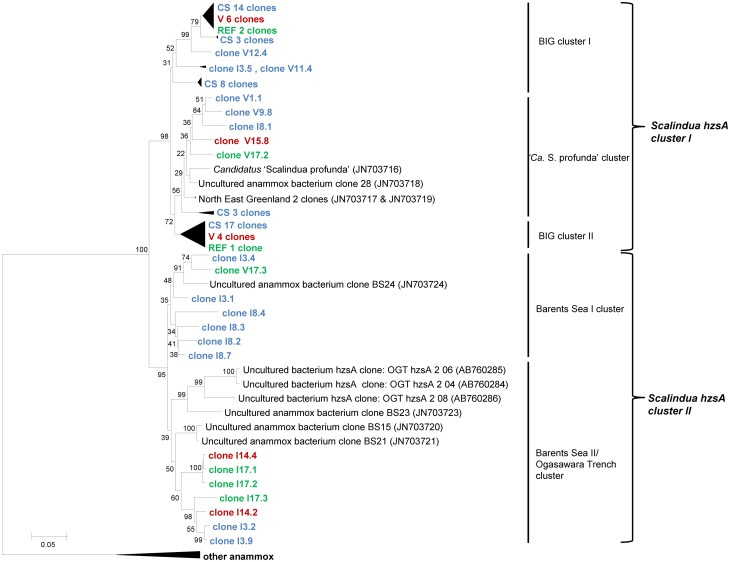
Figure 2.
Neighbor-joining tree of phylogeny estimated by ClustalW included in the MEGA 5.0 software package, showing >1000 bp fragments of hzsA nucleotide sequences retrieved from the Guaymas Basin sediments. Letters (I or V) of the samples indicate the primer set used for amplification and the number refers to the sample. Samples are color-coded and at collapsed nodes abbreviated: reference zone in green (REF), cold hydrocarbon-rich seeps in blue (CS) and hydrothermal sites in red (V). Values at the internal nodes indicate bootstrap values based on 500 iterations. The bar indicates 5% sequence divergence. The outgroup with other anammox bacteria includes Genbank accession numbers JN703715, JN703714, JN703713, JN703712, AB365070 and CT573071. Accession numbers of individual clones are provided in Supplementary Table 1.