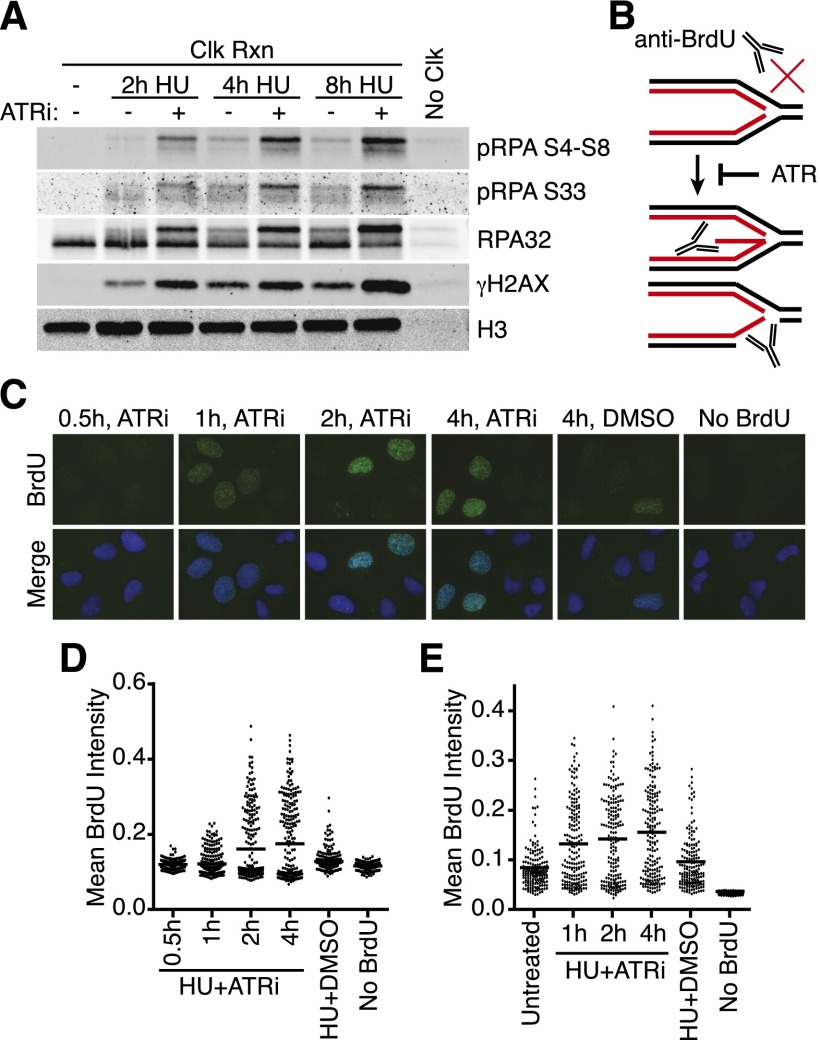
Figure 4.
ATR inhibition causes both nascent and parental ssDNA accumulation at stalled replication forks. (A) 293T cells were labeled with EdU for 10 min prior to addition of 3 mM HU and 5 μM ATRi as indicated. Samples were processed for iPOND, and captured proteins were separated by SDS-PAGE and then immunoblotted. (B) Model for nascent-strand ssDNA assay. Black and red lines indicate template and nascent DNA strands, respectively. Without DNA denaturation, BrdU antibodies will not recognize intact replication forks but will recognize the labeled, nascent DNA when single-stranded. (C,D) The newly synthesized DNA in replicating U2OS cells was labeled for 10 min with 10 μM BrdU before addition of 3 mM HU and 5 μM ATRi as indicated. “No BrdU” sample is 4-h HU+ATRi treatment without BrdU prelabeling. DMSO samples were labeled with BrdU and treated with 3 mM HU for 4 h. After the indicated treatment times, cells were fixed and stained with antibodies against BrdU without DNA denaturation to selectively detect nascent-strand ssDNA. Representative images are shown in C, and a dot plot of mean BrdU intensity per nucleus is shown in D. (E) Parental DNA in replicating U2OS cells was labeled by the addition of 10 μM BrdU for 20 h followed by a chase into normal growth medium for 2 h before addition of 3 mM HU and 5 μM ATRi for the indicated times. DMSO samples were labeled with BrdU and treated with 3 mM HU for 4 h. Cells were fixed and stained with antibodies against BrdU without DNA denaturation to selectively detect parental-strand ssDNA. Dot plot of mean BrdU intensity per nucleus is shown.