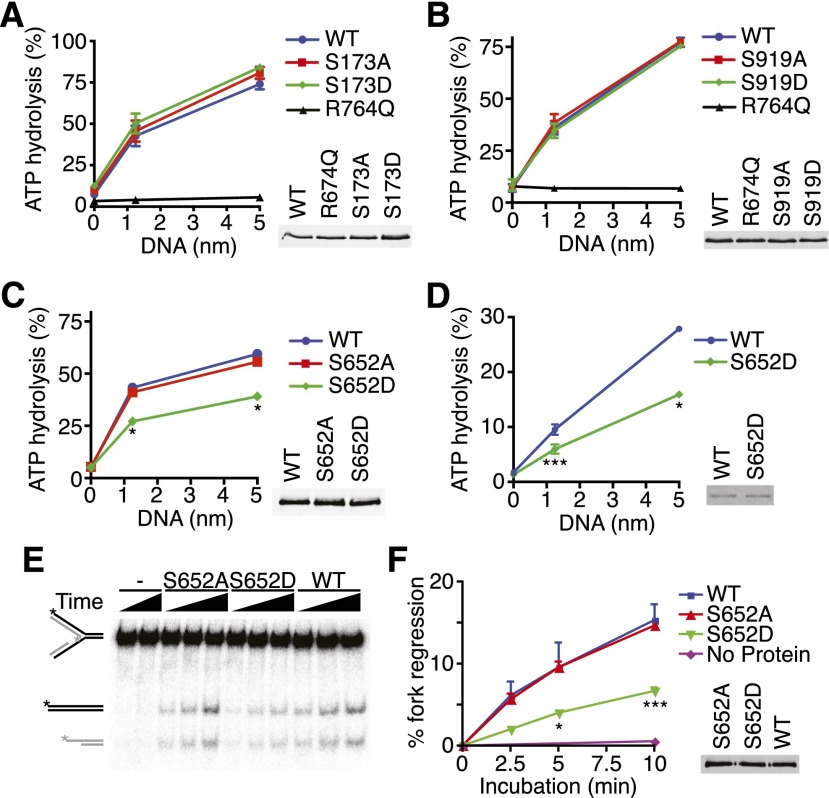
Figure 8.
SMARCAL1 phosphorylation on S652 inhibits its ATP-dependent fork remodeling activity. (A–D) The indicated Flag-SMARCAL1 proteins were purified from HEK293T cells (A–C) or baculovirus-infected insect cells (D), and their ATPase activity was measured in the presence of increasing concentrations of splayed arm DNA substrate. The insets in A–C are representative immunoblots, and the inset in D is a Coomassie-stained gel showing equal amounts of wild-type and mutant SMARCAL1 proteins used. Error bars in all panels represent SEM (n = 3) and in many cases were smaller than the symbol. (*) P < 0.0002; (**) P < 0.002; (***) P < 0.05. (E,F) The fork regression activity of purified SMARCAL1 proteins was assayed on a model replication fork substrate schematized on the far left. (See Supplemental Table S1 for details.) A representative DNA gel (E) and quantitation of three independent experiments (F) (mean and SEM) are shown. The inset is a representative immunoblot showing equal amounts of wild-type and mutant SMARCAL1 proteins.