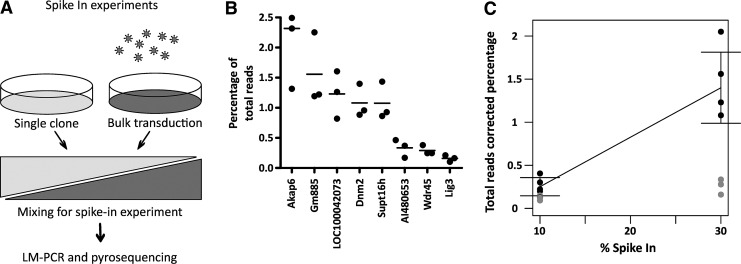
FIG. 4.
Quantification differences in a spike-in experiment. (A) The spike-in experiments were performed by mixing a clone with known vector insertions with a bulk transduced population, and the resulting mixture was analyzed by LM-PCR–pyrosequencing. (B) Percentage of total reads for the 30% spike-in sample, in triplicate, for the insertions in the clone, identified by the closest gene to the insertion. Bars indicate sample means. (C) Comparison of the percentage of total reads in the 10 and 30% spike-in samples. The insertions shown as solid circles demonstrate a clear increase in percentage of total reads, whereas the insertions shown as gray circles did not show a comparable increase.