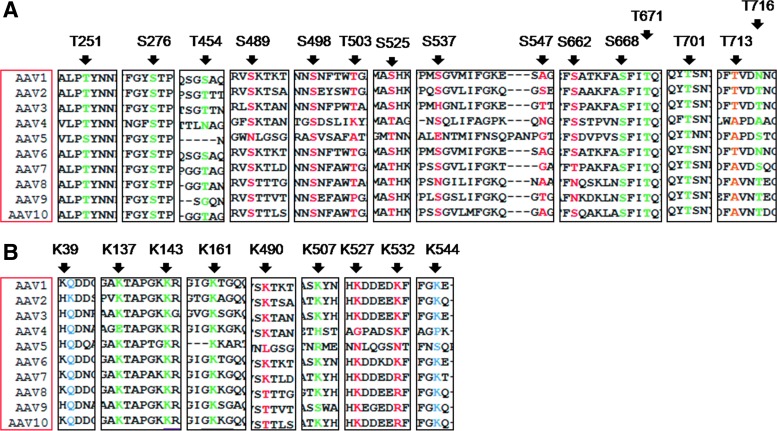
FIG. 2.
Schematic representation and conservation status of the various serine (S), threonine (T), and lysine (K) residues mutated in the AAV2 capsid. VP1 protein sequences from AAV serotypes 1 through 10 were aligned with ClustalW and the conservation status of each of the mutated sites is given. S/T residues are shown in (A) and lysine residues are shown in (B). S/T/K residues within phosphodegrons 1, 2, and 3 are shown in red whereas those chosen on the basis of evolutionary conservation are shown in green. Those residues that were chosen on the basis of either in silico prediction to be a part of a phosphosite or high ubiquitination score with the UbiPred tool are shown in blue. A control threonine mutation shown in brown was chosen as a negative control for the mutation experiments. Color images available online at www.liebertpub.com/hgtb