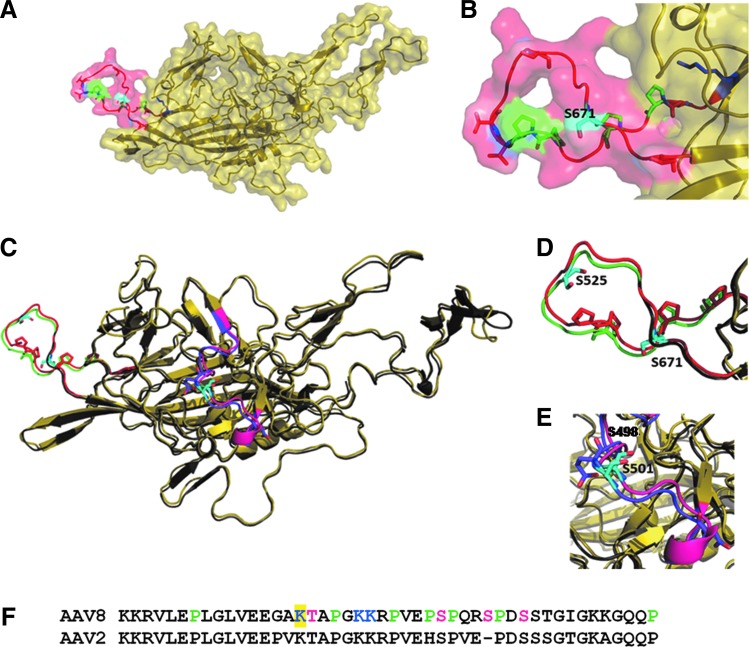
FIG. 2.
The phosphodegron in AAV8 capsid structure. (A) The figure shows the capsid protein from AAV8 (PDB id: 2qa0), which is colored yellow. S671 (colored cyan) lies in phosphodegron region (652–674), which is colored red. The phosphodegron is rich in prolines, which are colored green. The predicted phosphosites (serines and threonine) are shown in red while the predicted ubiquitination sites are in blue. (B) The zoomed-in view of phosphodegron region (652–674) containing S671. (C) Comparison of phosphodegrons in AAV8 and AAV2. The figure shows structural superimposition of AAV8 (PDB id: 2qa0) and AAV2 (PDB id: 1lp3) in yellow and gray, respectively. Phosphodegron in AAV8, colored red, is equivalent to phosphodegron2 in AAV2 (515–528), which is colored green. Phosphodegrons in both AAV2 and 8 are rich in proline residues. The residue S525 in AAV2 (colored cyan) lies in the phosphodegron and has been shown to increase the transduction efficiency (Gabriel et al., 2013). (D) The zoomed-in view of the phosphodegron2 in AAV2 and AAV8. (E) The zoomed-in view of phosphodegron3 of AAV2 in comparison with that in AAV8. The presence of another phosphodegron region in AAV2 (phosphodegron 3: residues 489-507), which is rich in acidic residues is colored purple. S489 and S498 in this region of AAV2 have shown to increase the transduction efficiency in vivo. Whereas, the equivalent region in AAV8, colored pink, lacks the acidic residues. (F) The figure shows the predicted phosphodegron stretch (122–137) in AAV8, which contains K137, highlighted in yellow. The phosphodegron is rich in proline and is colored green. The predicted phosphosites (T138, S149, S153, and S156) are colored magenta while the predicted ubiquitination sites (K137, K142, and K143) are colored blue. Color images available online at www.liebertpub.com/hgtb