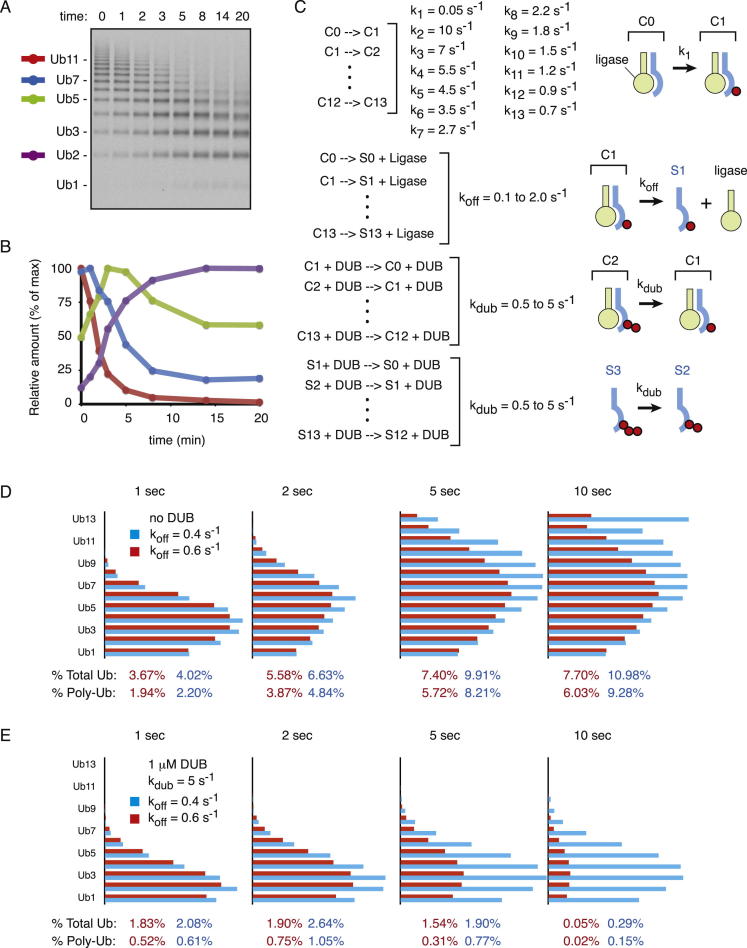
Figure S6.
Experimental and In Silico Analysis of DUB Activity, Related to Figure 5
(A) Deubiquitination of mCD4 by HEK293 cytosol. Proteoliposomes containing ubiquitinated mCD4 generated as in Figure S4B were isolated and incubated with HEK293 cytosol. Aliquots at different time points were analyzed by ubiquitin pulldowns and autoradiography.
(B) The Ub2 (i.e., substrate containing two ubiquitins), Ub5, Ub7, and Ub11 bands from A were quantified, normalized to maximal intensity of that species, and plotted. This analysis indicates that ubiquitins are successively removed from the distal ends of the polyubiquitin chains.
(C) The reactions used to model the ubiquitination and deubiquitination reactions illustrated in Figure 5B. In this model, C0 to C13 represent substrate-ligase complexes containing between 0 to 13 ubiquitins on the substrate. Hence, the reaction ‘C0 = C1’ represents a single ubiquitin addition. The kcat values for these reactions are indicated. The reactions of the type ‘C0 = S0 + Ligase’ are dissociation reactions of ligase from substrate (i.e., S0 to S13, depending on the number of ubiquitins attached). The deubiquitination reactions are allowed to occur on substrates that are either free or in a complex with ligase.
(D and E) Analysis of the normalized ubiquitin profiles generated by the model at different time points for the indicated parameters without or with DUB activity. The absolute amount of total and poly-ubiquitinated species at each time point is indicated below the respective graphs. Note that in the absence of DUBs (D), ubiquitination is very similar for the two substrates of differing Koff values. Only a modest difference is observed at later time points. By contrast, high DUB activity (E) results in a substantial (∼7-8 fold) difference in polyubiquitination over time. However, this comes at the cost of a marked loss in overall ubiquitination efficiency.