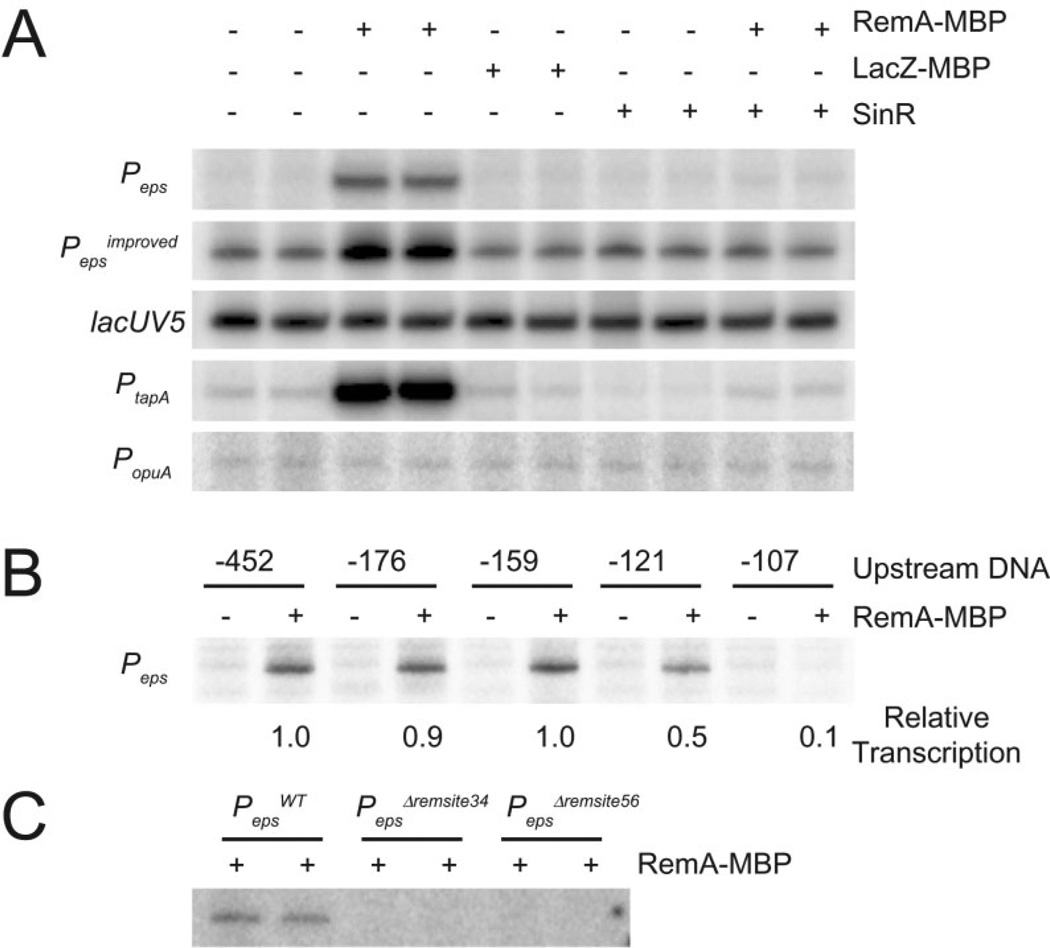
Fig. 2.
RemA activates transcription from specific promoters in vitro. In vitro transcription reactions were carried out in 100 mM KCl with 20 nM σA containing RNAP holoenzyme and 50 ng of supercoiled plasmid DNA with the indicated promoters cloned upstream of tandem transcription terminators.
A. RemA–MBP (700 nM), LacZ–MBP (700 nM) or SinR (400 nM) were added to in vitro transcription reactions containing the followingpromoters cloned upstream of tandem transcription terminators: Peps (−452 to +35), Pepsimproved (−452 to +35 with the −35 element changed from TTTAAA to TTGAAA), lacUV5 (−60 to +40), PtapA (−168 to +65) or PopuA (−173 to +26).
B. In vitro transcription reactions were carried out with supercoiled plasmid containing Peps promoter DNA with the indicated number of nucleotides upstream of the transcription start site cloned upstream of tandem transcription terminators. The downstream promoter end-point was +35 in each case. RemA–MBP (700 nM) or RemA–MBP storage buffer were added to each reaction. Relative transcription was determined by dividing the intensity in each lane by the intensity in lane 2. The indicated values for relative transcription are the averages from three different experiments.
C. In vitro transcription reactions were carried out with supercoiled plasmid containing 700 nM RemA–MBP and either WT Peps (−452 to +35)or Peps promoter fragments containing the indicated Δremsite34 or Δremsite56 deletions.