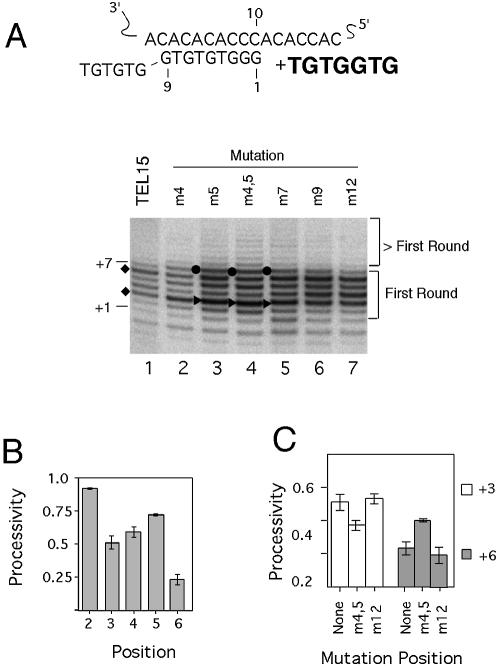
Figure 2.
Effects of primer mutations on yeast telomerase elongation. (A) An alignment of the template region of yeast telomerase RNA with the 15-nt canonical yeast primer (TEL15) is shown at the top. The template residues and the primer residues are both numbered from 3′ to 5′. The 3′ most residue of the primer should be positioned opposite the 10th nucleotide of the template. The sequence that is expected to be added by telomerase is shown in bold. Results of primer extension assays that were performed using TEL15 and related primers, 0.2 µM dGTP and 50 µM dTTP are shown in the bottom panel. The positions of the mutations (either T to A or G to C) in the DNA primers are indicated at the top of the panel. The locations of the P + 1 and P + 7 products are indicated at the left side of the panel. The diamonds point to the locations of the two elongation barriers for TEL15, at the P + 3 and P + 6 positions. The filled triangles and circles point to changes in the relative intensities of the P + 3 and P + 7 products, respectively. (B) The processivity of telomerase for the TEL15 primer from the P + 2 to P + 6 positions are plotted. (C) The processivity of telomerase for the TEL15, TEL15(m4,5) and TEL15(m12) primers at the P + 3 and P + 6 positions are plotted.