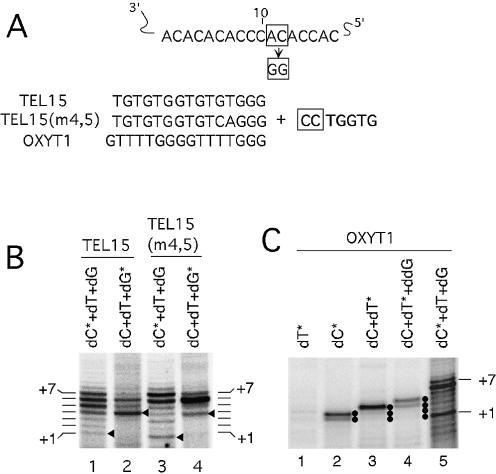
Figure 7.
The primers align to the RNA template in the predicted manner. (A) The predicted alignment of the template region of the mutated yeast telomerase RNA with the 15-nt yeast primers [TEL15 and TEL15(m4,5)] and a heterologous primer that also ends in 3Gs (OXYT1) is shown. The template residues are numbered from 3′ to 5′. The 3′ most residue of the primers are predicted to be positioned opposite the 10th nucleotide of the template. The sequence that is expected to be added by the mutant telomerase is also shown with the first two dC residues boxed. (B) Primer extension assays were performed using either TEL15 (lanes 1 and 2) or TEL15(m4,5) (lanes 3 and 4) and telomerase that was purified by DEAE chromatography. All reactions contain the three nucleotides that are necessary for copying the entire mutant template (dCTP, dGTP and dTTP). The labeled nucleotides (indicated by asterisks) and the unlabeled nucleotides are present at 0.2 and 50 µM, respectively. The positions of the P + 1 to P + 7 products are indicated at both sides of the panel. (C) Primer extension assays were performed using OXYT1 as primer, different combinations of labeled nucleotides (specified by asterisks) and unlabeled nucleotides as indicated at the top, and telomerase that was purified by DEAE chromatography. The positions of the P + 1 and P + 7 products are indicated on the right side.