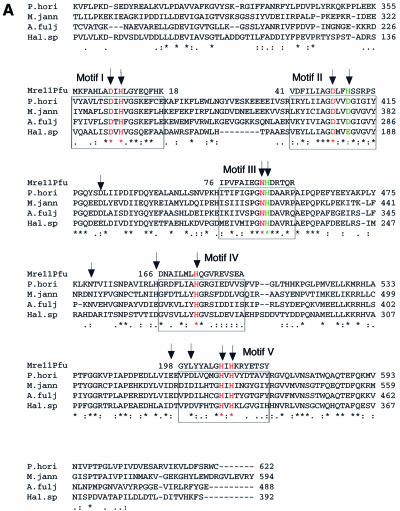
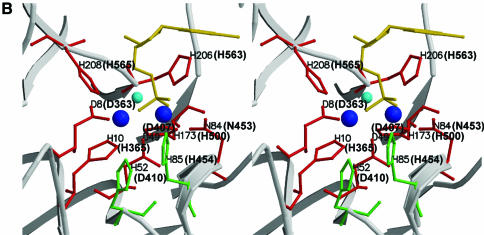
Figure 1.
Site-directed mutation sites at PD1Pho in the PolD complex. (A) Partial sequence alignment of the DP1s from Euryarchaeota species. The DP1 sequences are from four Euryarchaeota species, P.horikoshii (P.hori, accession number, AP000001–127), Methanococcus jannaschii (M.jann, U67516–16), Archaeoglobus fuljidus (A.fulj, AE000979–13) and Halobacterium sp. NRC-1 (Hal.sp, AE005122–10). The alignment was carried out using ClustalW software. The motifs (I–V) conserved among the phosphoesterase super-family are boxed. The corresponding motifs from Mre11 of P.furiosus (Mre11Pfu) are shown above the boxes. The residues colored in red are putatively involved in metal binding and those in green are putative catalytic assistance residues according to the structure of Mre11 from P.furiosus. The arrows indicate residues chosen for the site-directed mutagenesis in this study. (B) Stereoview showing the catalytic site of Mre11Pfu. Structural data (1II7, ref. 37) were retrieved from the Protein Data Bank and the figure was made with Molscript (51) and Raster3D (52). The residues involved in metal binding are shown in red and those putatively involved in catalytic assistance in green. The bound dAMP is shown in yellow. The residues of Mre11Pfu are labeled together with their corresponding residues of DP1Pho (in brackets). Two manganese ions and a water molecule are shown in blue and cyan respectively.