Abstract
The Fenna-Matthews-Olson antenna protein from the green bacterium Pelodictyon phaeum mediates the energy transfer from a peripheral antenna complex to the membrane-bound reaction center. The three-dimensional structure of this protein has been previously modeled using X-ray diffraction to a resolution limit of 2.0 Å, with Rwork and Rfree values of 16.6% and 19.9% respectively (Larson et al. (2011) Photosyn Res 107: 139–150). This model shows the protein as consisting of β-sheets surrounding several bacteriochlorophyll cofactors. While most of the model clearly matches the electron density maps, in this paper we re-examine the electron density for a specific feature, namely the eighth bacteriochlorophyll a cofactor. This electron density is now interpreted as arising primarily from the end of an otherwise disordered polyethylene glycol molecule. Additional electron density is present but the density is weak and cannot be unambiguously assigned. The new model has Rwork and Rfree values of 16.2% and 19.0%, respectively.
Keywords: Energy transfer, light harvesting complex, three dimensional structure, green bacteria
Introduction
Many different types of bacteriochlorophyll-containing proteins are utilized by photosynthetic organisms to convert light energy into chemical energy (Blankenship 2002). In green sulfur bacteria, light is initially absorbed by large complexes termed chlorosomes (Staehelin et al. 1980; Psencik et al. 2004; Oostregetel et al. 2007) and transferred to a bacteriochlorophyll a (Bchl-a) protein termed the Fenna-Matthews-Olson (FMO) protein (Olson 2004). The FMO protein directs the energy to the membrane-embedded reaction center that is the site for the energy conversion (Wen et al. 2009). The ability of the Bchl-a cofactors to perform energy transfer has been extensively characterized using optical spectroscopy, including time resolved two-dimensional spectroscopy that allowed a molecular interpretation of energy flow and energy couplings within the FMO protein (Brixner et al. 2005; Read et al. 2007; Panitchayangkoon et al. 2010; Milder et al. 2010)
Key to the interpretation of these spectroscopic studies is the availability of the three-dimensional structures of three FMO proteins. The FMO complex from Prosthecochloris aestuarii 2K (Ptc. aestuarii 2K) was the first Bchl-a containing protein to be crystallized and have its three dimensional structure determined (Olson 1978; Matthews et al. 1979; Tronrud et al. 1986; Tronrud et al. 2009). The structure of the FMO complex from Chlorobaculum tepidum (Cbl. tepidum), previously named Chlorobium tepidum, was then determined followed by the FMO protein from Pelodictyon phaeum (Pld. phaeum) (PDB code 3OEG) (Li et al. 1997; Camara-Artigas et al. 2003; Ben-Shem et al. 2004; Tronrud et al. 2009; Larson et al. 2011). These structures show that the FMO protein is a complex with three identical subunits related by a 3-fold symmetry axis, thus forming a homotrimer. Each subunit forms two large β-sheets surrounding seven Bchl-a cofactors that are all buried within the protein.
A surprising feature of some recent structures was the presence of an eighth Bchl-a molecule located the protein-protein interface of the trimer (Ben-Shem et al. 2004; Tronrud et al. 2009). Unlike the other Bchl-a molecules, Bchl-a 8 is not buried in the protein but rather is located at a cleft at the interface between protein subunits forming the trimer. It was suggested that the lack of electron density for Bchl-a 8 for the previous work on the FMO from Cbl. tepidum was due to a change in the isolation of the protein suggesting that Bchl-a 8 was more labile than the other seven (Ben-Shem et al. 2004). Examination of a high resolution data set of crystals of the FMO from Ptc. aestuarii 2K also showed the presence of Bchl-a 8 although the occupancy was very low (≈ 30 %) (Tronrud et al. 2009). Diffraction data from FMO from Cbl. tepidum was re-examined and a model was created with the constraint of a fully occupied Bchl-a 8 even though the occupancy was actually less than unity (Tronrud et al. 2009). For the structure of the FMO from Pld. phaeum, a Bchl-a molecule was placed at the corresponding site based upon significant electron density in omit maps (Larson et al. 2011). However, the refined location of this Bchl-a molecule was shifted relative to the other structures resulting in different interactions within the binding pocket. The molecule did not penetrate as deeply into the binding pocket and its magnesium atom was linked only to the side chain of serine 164. Also, while the electron density maps covered much of the Bchl-a 8, the electron density was weak over some portions, which was interpreted as arising due to a disorder for part of the cofactor (Larson et al. 2011). The model of this Bchl-a molecule had very poor agreement with its expected planarity and there were several very short contacts between the Bchl-a and neighboring protein atoms. This model contradicted a correlation evident between a comparison of FMO sequences from different organisms and the identity of the amino acid residues forming the binding site, in particular the amino acid residue coordinating the central magnesium atom (Tronrud et al. 2009).
In this paper, the electron density for this region is re-interpreted and the resulting new model of the FMO from Pld. phaeum is described.
Materials and methods
The protein crystallization and diffraction measurements have been previously described (Larson et al. 2011). That model has all of the amino acid residues except for the first four, N-terminal, residues and two residues, 209 and 210, that are located in a loop and disordered. The structure was built using Coot (Emsley and Cowtan 2004) and refined using PHENIX (Adams et al. 2002). All residues were in either the preferred or allowed regions of the Ramachandran plot, and the completed model had Rwork and Rfree values of 16.6% and 19.9% respectively. The new model was refined using Buster (Bricogne et al. 2009) and manually refit using Coot. Three rounds were performed in which three additional fragments of PEG were built that bind to other parts of the FMO protein and a number of small corrections were made. The final Rwork and Rfree values are 16.21% and 19.02%, respectively. The new model has been deposited in the PDB with the code 3VDI and 3OEG has been flagged as obsolete.
Results and Discussion
The structure of the FMO from Pld. phaeum shows the same dominant conserved features seen in the models of FMO from Ptc. aestuarii 2K and Cbl. tepidum, namely a β-fold that surrounds seven Bchl-a cofactors. Aside from minor adjustments, these features of the model are not significantly altered in the newly refined model and will not be discussed further. Rather, the focus here is on the interpretation of the electron density in the region of BChl-a 8.
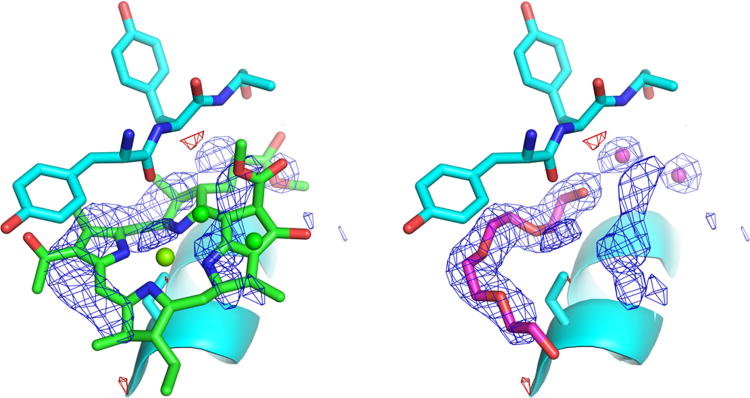
Figure 1 shows the omit electron density map calculated for the binding site of BChl-a 8. The omit density is composed of two tubular objects along with several small globular peaks. The most electron dense of the tubes (on the left of the figure) wraps around the carbonyl oxygen of residue 119. The weaker tube of density is not close enough to the protein to have direct hydrogen bonds or van der Waal interactions. There are several spherical bits of density which have shapes consistent with that of water molecules and a pair them (in the distance, on the right) have distances to other objects that are consistent with hydrogen bonds.
Fig 1.
Superposition of each model with the omit electron density map for the binding pocket. The map was calculated by removing the Bchl-a molecule and water molecules from this binding site in PDB model 3OEG and refining the result with the Buster software package (Bricogne et al. 2009). The same map is shown in both panels and was contoured at ±3 times the root mean square of the overall density. a) The Bchl-a 8 and two water molecules from the published model (3OEG). b) The new model (3VDI) with its PEG fragment and two water molecules occupying the strongest density features. (Figure created with the PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.)
Since the electron density observed in this binding pocket is not planar, refinement of the Bchl-a modeled in 3OEG resulted in severe distortions. Its macrocycle has been folded between rings I and IV to minimize its clash with the carbonyl oxygen of residue 119. The refinement program tried to balance the consequences of these bad contacts by distorting the planarity of the macrocycle but in the end neither restraint was satisfied. The worst of the bad contacts is to the NA atom in ring IV and is 2.94 Å long. (This contact cannot be a hydrogen bond because neither of these atoms is expected to have a hydrogen atom and the geometry of the interaction is highly unfavorable.)
In addition, the model for the Bchl-a molecule is not centered in the tubular electron density. From the point of view of Figure 1a, the entire molecule must be shifted to the left. It was not modeled this way because the COCH3 substituent of ring I would then be forced too close to the protein atoms that form the wall of the pocket. The result is the presence of large peaks in the final difference electron density map for 3OEG.
For the rest of the Bchl-a molecule the electron density is either weak or nonexistent. The density is broken between rings V and IV, and ring II has no density at all. The magnesium atom, the only atom with a proposed contact to the protein, lies outside of electron density.
A new model of the contents of this pocket has been built that fits the electron density well and agrees with stereochemical restraints. The principal tube of electron density is fit with a fragment of polyethylene glycol (PEG). The small globular pieces of density are modeled with water molecules. The mother liquor for this crystal contained polyethylene glycol 2000 monomethyl ether (Larson et al. 2011). The new model has excellent stereochemistry, a good hydrogen bonding network, and no bad contacts. The quality of the fit to the original omit map is shown in Figure 1b.
The second tubular density cannot be modeled with confidence. The density is weaker than its surroundings and the molecules contributing to this electron density are likely to have significant disorder. This density may arise from a second PEG fragment, or a Bchl-a molecule with low occupancy. However, the presence of other molecules is also possible and so the electron density in this region has been left uninterpreted in the revised model. Regardless of what is binding at this location, we are confident that the primary density in this pocket reflects the binding of a PEG fragment. Since this structural model does not contain Bchl-a 8, the accuracy of the predictions in Tronrud et al. (2009) remains an open question. This outcome also suggests that the low occupancy of Bchl-a 8 in the crystals of FMO from other species probably arises due to loss during the purification and crystallization of the protein with the functional state being fully occupied.
Abbreviations
- FMO
Fenna-Matthews-Olson
- Bchl-a
bacteriochlorophyll a
- PEG
polyethylene glycol
Contributor Information
Dale E. Tronrud, Email: tronrudd@science.oregonstate.edu.
James P. Allen, Email: JAllen@asu.edu.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Mariarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N. Evolution of photosystem I-from symmetry through pseudosymmetry to asymmetry. FEBS Lett. 2004;564:274–280. doi: 10.1016/S0014-5793(04)00360-6. [DOI] [PubMed] [Google Scholar]
- Blankenship RE. Molecular Mechanisms of Photosynthesis. Wiley-Blackwell; Malden MA: 2002. [Google Scholar]
- Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Smart OS, Vonrhein C, Womack TO. Buster, version 2.8.0. Global Phasing Ltd.; Cambridge, U.K: 2009. [Google Scholar]
- Brixner T, Stenger J, Vaswani HM, Cho M, Blankenship RE, Fleming GR. Two-dimensional spectroscopy of electronic couplings in photosynthesis. Nature. 2005;434:625–628. doi: 10.1038/nature03429. [DOI] [PubMed] [Google Scholar]
- Camara-Artigas A, Blankenship RE, Allen JP. The structure of the FMO protein from Chlorobium tepidum at 2.2 Å resolution. Photosynth Res. 2003;75:49–55. doi: 10.1023/A:1022406703110. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Larson CR, Seng CO, Lauman L, Matthies HJ, Wen J, Blankenship RE, Allen JP. The three dimensional structure of the FMO protein from Pelodictyon phaeum and the implications for energy transfer. Photosyn Res. 2011;107:139–150. doi: 10.1007/s11120-010-9604-2. [DOI] [PubMed] [Google Scholar]
- Li YF, Zhou W, Blankenship RE, Allen JP. Crystal structure of the bacteriochlorophyll a protein from Chlorobium tepidum. J Mol Biol. 1997;271:456–471. doi: 10.1006/jmbi.1997.1189. [DOI] [PubMed] [Google Scholar]
- Matthews BW, Fenna RE, Bolognesi MC, Schmid MF, Olson JM. Structure of a bacteriochlorophyll a-protein from the green photosynthetic bacterium Prosthecochloris aestuarii. J Mol Biol. 1979;131:259–285. doi: 10.1016/0022-2836(79)90076-7. [DOI] [PubMed] [Google Scholar]
- Milder MTW, Brüggemann B, van Grondelle R, Herek JL. Revisiting the optical properties of the FMO protein. Photosynth Res. 2010;104:257–274. doi: 10.1007/s11120-010-9540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JM. Bacteriochlorophyll a-proteins from green bacteria. In: Clayton RK, Sistrom WR, editors. The Photosynthetic Bacteria. Plenum Press; NY: 1978. pp. 161–178. [Google Scholar]
- Olson JM. The FMO protein. Photosynth Res. 2004;80:181–187. doi: 10.1023/B:PRES.0000030428.36950.43. [DOI] [PubMed] [Google Scholar]
- Oostergetel GT, Reus M, Chew AGM, Bryant DA, Boekema EJ, Holzwarth AR. Long-range organization by bacteriochlorophyll in chlorosomes of Chlorobium tepidum investigated by cryo-electron microscopy. FEBS Lett. 2007;581:5435–5439. doi: 10.1016/j.febslet.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Panitchayangkoon G, Hayes D, Fransted KA, Caram JR, Harel E, Wen J, Blankenship RE, Engel GS. Long-lived quantum coherence in photosynthetic complexes at physiological temperature. Proc Natl Acad Sci USA. 2010;107:12766–12770. doi: 10.1073/pnas.1005484107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psencik J, Ikonen TP, Laurinmäki P, Merckel MC, Butcher SJ, Serimaa RE, Tuma R. Lamella organization of pigments in chlorosomes, the light harvesting complexes of green photosynthetic bacteria. Biophys J. 2004;87:1165–1172. doi: 10.1529/biophysj.104.040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read EL, Engel GS, Calhoun TR, Mančal T, Ahn TK, Blankenship RE, Fleming GR. Cross-peak-specific two-dimensional electronic spectroscopy. Proc Natl Acad Sci USA. 2007;104:14203–14208. doi: 10.1073/pnas.0701201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA, Golecki JR, Drews G. Supermolecular organization of chlorosomes (Chlorobium vesicles) and their membrane attachment sites in Chlorobium limicola. Biochim Biophys Acta. 1980;589:30–45. doi: 10.1016/0005-2728(80)90130-9. [DOI] [PubMed] [Google Scholar]
- Tronrud DE, Schmid MF, Matthews BW. Structure and X-ray amino acid sequence of a bacteriochlorophyll a protein from Prosthecochloris aestuarii at 1.9 Å resolution. J Mol Biol. 1986;188:443–454. doi: 10.1016/0022-2836(86)90167-1. [DOI] [PubMed] [Google Scholar]
- Tronrud DE, Wen J, Gay L, Blankenship RE. The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria. Photosynth Res. 2009;100:79–87. doi: 10.1007/s11120-009-9430-6. [DOI] [PubMed] [Google Scholar]
- Wen J, Zhang H, Gross ML, Blankenship RE. Membrane orientation of the FMO antenna protein from Chlorobaculum tepidum as determined by mass spectroscopy-based footprinting. Proc Natl Acad Sci USA. 2009;106:6134–6139. doi: 10.1073/pnas.0901691106. [DOI] [PMC free article] [PubMed] [Google Scholar]