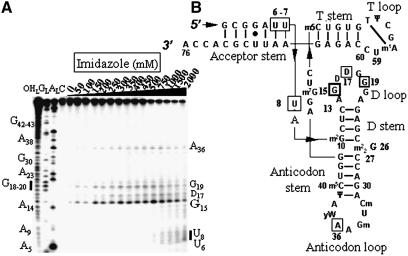
Figure 2.
Electrophoregram of the cleavage pattern of labeled tRNAPhe by an imidazole buffer. (A) Lane C, incubation control; lane OHL, alkaline hydrolysis ladder; lane GL, guanine ladder; lane AL, adenine ladder. Sequence is indexed on the left side. Nucleotides indexed on the right side are the cleavage sites. (B) Yeast tRNAPhe secondary structure with imidazole induced cleavage points. Thickness of the symbols (rectangles) is proportional to the intensity of the cuts. Nucleotides at the 5′-boundary of the cleavage sites are indicated onto tRNAPhe secondary structure, the cleavage sites are at the 3′-phosphate. This molecule has 11 modified bases: N2-methylguanosine (m2G), dihydrouridine (D), N2,N2-dimethylguanosine (m22G), 2′-O-methylcytidine (Cm), 2′-O-methylguanosine (Gm), wybutosine (yW), pseudouridine (Ψ), 5-methylcytidine (m5C), 7-methylguanosine (m7G), 5-methyluridine (m5U) and 1-methyladenosine (m1A).