Abstract
Dysregulation of proteolytic processing of the amyloid precursor protein (APP) contributes to the pathogenesis of Alzheimer’s Disease, and the Group VIA phospholipase A2 (iPLA2β) is the dominant PLA2 enzyme in the central nervous system and is subject to regulatory proteolytic processing. We have identified novel N-terminal variants of iPLA2β and previously unrecognized proteolysis sites in APP constructs with a C-terminal 6-myc tag by automated identification of signature peptides in LC/MS/MS analyses of proteolytic digests. We have developed a Signature-Discovery (SD) program to characterize protein isoforms by identifying signature peptides that arise from proteolytic processing in vivo. This program analyzes MS/MS data from LC analyses of proteolytic digests of protein mixtures that can include incompletely resolved components in biological samples. This reduces requirements for purification and thereby minimizes artifactual modifications during sample processing. A new algorithm to generate the theoretical signature peptide set and to calculate similarity scores between predicted and observed mass spectra has been tested and optimized with model proteins. The program has been applied to the identification of variants of proteins of biological interest, including APP cleavage products and iPLA2β, and such applications demonstrate the utility of this approach.
Deposition of amyloid in central nervous system tissue is a key event in the pathogenesis of Alzheimer’s Disease, and peptides derived from cleavage of the amyloid precursor protein (APP) differ in their amyloidogenicity [1, 2]. Considerable effort is thus directed at characterizing the proteolytic processing of APP by identifying the responsible proteases, their cleavage sites, and the resultant product peptides. The Group VIA Phospholipase A2 (iPLA2β) is the dominant cytosolic PLA2 activity in the central nervous system, and a number of isoforms of this enzyme with different properties are expressed in various cells [3–10]. Some iPLA2β variants arise from alternative splicing of transcripts, and others arise from proteolytic processing at both the C-terminus and the N-terminus of the parent iPLA2β sequence. Characterization of cleavage sites of iPLA2β is required to understand its processing and the structures and functions of its variant isoforms.
Identification of variants of proteins that arise from the same gene, such as that for APP or iPLA2β, is a problem in proteomics [11], which is the study of gene products that are far more diverse than the set of genes from which they arise. Among mechanisms for generating such diversity are alternative splicing of transcripts and post-translational modification (PTM) [12–14]. PTM can include covalent modification of amino acid residues, e.g., phosphorylation of side-chain hydroxyl groups, or proteolytic processing by exopetidases or endopeptidases. Mass spectrometry has now become a principal tool for identification of proteins and their variant forms [11]. Fairly pure proteins with MW below 10 kDa can often be successfully analyzed by MALDI/TOF/MS. Accurate mass measurements facilitate identification of protein variants or isoforms with high throughput [15], but the MW of many proteins is greater than 10 kDa. Proteins in biologic samples are also often incompletely resolved components of complex mixtures, and accurate mass measurements alone may be insufficient to achieve unambiguous identifications of protein variants or isoforms.
A frequently employed approach to protein identification is to digest the sample with proteolytic enzymes of known sequence specificity [11]. For a purified sample containing a small number of proteins, peptide mapping is often sufficient to identify proteins by comparing the observed proteolytic peptide masses with the predicted peptide masses calculated from protein or gene databases that specify protein sequences directly or by deduction [16–22]. For more complex protein mixtures, tandem mass spectrometry (MS/MS) is required to obtain amino acid sequence information, which is deduced from fragmentation patterns of individual proteolytic peptides [11]. Computer assistance is required to process the large amount of data generated by LC/MS/MS analyses of such peptide mixtures. The MS/MS data are submitted to database searching [23–25] or to de novo sequencing programs [26–28]. Some programmed algorithms have been developed to identify protein PTMs [29, 30] from MS/MS data.
When a protein in a biological sample represents the product of in vivo proteolytic processing of a parent protein, digestion of the product protein in vitro with exogenous proteases, e.g., trypsin, to yield a set of peptides for protein identification often generates a signature peptide that can be used to identify the site of in vivo proteolytic processing. A signature peptide is one that arises from digestion of a protein derived from the same gene as a parent protein, but which lacks a region of amino acid sequence contained by the parent protein. Such peptides would not be produced from digestion of the parent protein by the given protease but appear because of missing sequence in the protein variant that has been removed by in vivo processing.
An example of identifying a signature peptide that reflects an in vivo proteolytic processing event appeared in one of the earliest reports of what has become a standard approach in proteomics, which is to analyze proteins from a biological source by 2-D gel electrophoresis, to subject isolated protein spots to tryptic proteolysis, and to analyze the peptides in the digest by peptide mass mapping with MALDI/TOF/MS and then, when necessary, by LC/MS/MS to obtain peptide sequence information [31]. When analyzing a yeast proteome in this manner, Shevchenko et al. encountered a protein of apparent molecular mass of 38.8 kDa and isoelectric point of 6.45 that yielded a peptide mass map that had 30 peaks that closely matched expected tryptic peptides for yeast protein ILV5, but native ILV5 has a calculated molecular mass of 44.4 kDa and an isoelectric point of 9.46 [31].
Because the C-terminal tryptic peptide of ILV5 expected from the native sequence was observed in the MALDI/TOF mass spectrum, it was suspected the 38.8 kDa protein arose from 44.4 kDa ILV5 by N-terminal proteolytic processing. The N-terminal sequence of ILV5 is Arg/Lys-rich, and removal of residues 1–47 from the 44.4 kDa sequence would yield a protein of 38.8 kDa with an isoelectric point of 6.45. Because the peptide bond between residues 47 and 48 does not represent a normal tryptic cleavage site, digestion of the 38.8 kDa protein with trypsin would be expected to yield a new tryptic peptide beginning at residue 48 with an [M + H]+ m/z value of 1853.909. Such a peak was observed in the MALDI/TOF mass spectrum, confirming that the 38.8 kDa protein arose from removal of residues 1–47 of the native 44.4 kDa ILV5 sequence. The peptide of m/z 1853.909 is thus a “signature peptide” that reflects nontryptic proteolytic processing that has presumably occurred in vivo [31].
Sequence information can also be useful in identifying such signature peptides. Figure 1 illustrates a case that will be described further in the Results and Discussion section. For the iPLA2β enzyme, the cloned cDNA encodes a protein with the sequence 1MGFFGR…. Digestion of the native sequence with trypsin would be expected to yield the peptides 1M-R6, 7L-R23, 24A-K25, 26EVSLA…, but iPLA2β is subject to N-terminal proteolytic processing. Among the variants produced is one from which residues 1M-L11 have been removed to yield a truncated protein that begins with residue 12S. Tryptic digestion of this truncated variant yields the peptide 12S-R23, which would not be expected to arise from tryptic digestion of the full-length protein. The sequence of the peptide 12S-R23, however, matches that encoded by iPLA2 cDNA. In this case, the peptide 12S-R23 is a signature peptide that reflects nontryptic proteolytic processing to remove the N-terminal 11 amino acid residues from the full-length iPLA2β sequence [3].
Figure 1.
Illustration of a signature peptide from an iPLA2β variant that reflects in vivo proteolytic processing.
To identify such peptides, derivatization methods have been used to isolate N-terminal signature peptides from digest mixtures [32]. Although derivatization is sometimes incomplete and digestion with a single protease might yield low sequence coverage, this approach has achieved identification of 264 proteins from human thrombocytes [32]. Eleven of them were different isoforms of the same gene product that were produced by N-terminal processing. Because de novo sequencing has limited accuracy and because peptide mixtures that result from digestion of incompletely purified biological samples are complex, it is often difficult to identify signature peptides by direct inspection of the MS/MS data, and, computer assistance with data interpretation is required.
Here we describe an approach to analyze LC/MS/MS data from peptide mixtures that can identify signature peptides and protein isoforms that arise from endogenous proteolytic processing. A new method to generate the signature peptide candidates has been developed for our approach and greatly reduces the number of members of the signature peptide candidate set and thereby reduces computational time. In addition, we have developed an optimized similarity score calculation that considers both fragmentation and intensity information, making the signature peptide identification more reliable. Our Signature-Discovery (SD) program implements this approach to analyze LC/MS/MS data of proteolytic digests of protein mixtures and identify signature peptides automatically. The SD program performance has been tested with model proteins and biological samples from cellular expression systems. Such analyses have resulted in identification of previously unrecognized cleavage sites for processing of iPLA2β and of an APP construct.
Experimental Procedures
Materials
All chemicals were purchased from Sigma Chemical (St. Louis, MO) and all solvents from Fisher Chemical (St. Louis, MO) unless otherwise stated. PepMap HPLC columns and pre-columns were obtained from LC-Packings (San Francisco, CA).
Cloning, Expression, and Purification of Native and His-Tagged iPLA2β Proteins in Sf9 cells
S. frugiperda (Sf9) cells were cultured as described elsewhere [33–36]. For expression of iPLA2β protein, a 250-ml flask was prepared with a suspension of 50 ml of 106 cells/ml and incubated at 27°C for 24 h before infection with baculovirus. For protein expression, cDNA containing the entire coding sequence of native iPLA2β [37] or his-tagged iPLA2β with the tag at either the C-terminus or at the N-terminus [38], or was cloned into the EcoRI-SalI site of the pFASTBACtm 1 baculovirus expression vector. The sequence of the insert was verified, and the plasmid was then transformed into DH10Bac cells according to instructions in the Bac-To-Bac system manual. Recombinant bacmid DNA was isolated from small-scale cultures using an alkaline lysis protocol modified for high-molecular-weight plasmid purification. PCR analysis was performed with purified bacmid DNA and pUC/M13 amplification primers to determine the size of inserts.
Typically, Sf9 cell suspensions (50 ml of 106 cells/ml) in supplemented Grace’s medium were infected with 1 ml of baculovirus. Sf9 cells were collected by centrifugation (500 × g, 10 min, 4°C), washed with PBS buffer, and re-suspended in lysis buffer (25 mM NaH2PO4, 2 mM β-mercaptoethanol, 5 μg/ml leupeptin, pH 7.8). The cells were disrupted by sonication, and the cytosol was prepared by subsequent centrifugation (18,000 × g, 15 min). His-tagged proteins were purified with a TALON metal affinity column and detected by Coomassie stain protein gel and immunoblotting. The Sf9 cell homogenate containing his-tagged iPLA2β proteins was mixed with TALON metal affinity resin (Clontech, Palo Alto, CA) and incubated at 4°C with shaking for 1 h. The mixture was washed with 10 bed volumes of wash buffer (50 mM Na2HPO4, 500 mM NaCl, 10 mM imidazole, pH 7.8) twice and transferred onto a gravity-flow column. The his-tagged iPLA2β was eluted with elution buffer (50 mM Na2HPO4, 300 mM NaCl, and 200 mM imidazole, pH 7.8) and collected in 0.5 ml fractions. Native iPLA2β was purified as described elsewhere [9, 34, 35]. Coomassie stained protein gels and immunoblotting analyses were used to visualize iPLA2β and his-tagged iPLA2β.
Generation and Immunoprecipitation of Proteolytic Products from myc-Tagged APP Constructs
The cDNA constructs for presenilins and APP-6myc were generated as described previously [39]. Human embryonic kidney 292 (HEK) cells were obtained from the ATCC (Rockville, MD) and maintained in DMEM (GibcoBRL, Grand Island, NY) containing 10% fetal bovine serum (GibcoBRL), 2 mM L-glutamine, 100 μg/ml penicillin under a 5% CO2/95% air atmosphere. Pooled stable cell lines expressing presenilins were generated by transfecting HEK parental cells with the appropriate DNA constructs and selecting with G418, as previously described [39]. Cells were transfected with various constructs using FuGENE 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN). Forty-eight h after transfection, cells were lysed in co-IP lysis buffer (50 mM Tris, pH 7.6, 150 mM NaCl, 2 mM EDTA, 1% NP40 and 0.5% Triton X-100). About 10% of the lysates were used for immunoblotting analysis. The remaining lysates were cleared with Protein A agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) and incubated with anti-myc monoclonal antibody 9E10 (Sigma) and 40 μl Protein A at 4°C overnight with constant rotation. The immunoprecipitates were washed twice with wash buffer 1 (50 mM Tris, pH 7.6, 500 mM NaCl, and 2 mM EDTA), twice with wash buffer 2 (50 mM Tris, pH 7.6, 150 mM NaCl, 2 mM EDTA), as previously described [39], and then suspended in buffer for proteolysis.
Digestion with Proteolytic Enzymes
Samples were concentrated to dryness under nitrogen and rehydrated in a solution of 50 mM NH4HCO3. Sequencing grade modified trypsin (Promega Corp. Madison, WI) or sequencing grade modified Glutamic C endopeptidase (Princeton Separations, Adelphia, NJ) was added at a ratio of 1/50 (wt/wt). After 12 h incubation at 37°C, the digest was adjusted to pH 2 and analyzed by LC/MS/MS.
MALDI/TOF Mass Spectrometry
MALDI/TOF mass spectra were acquired with a Voyager DE STR instrument (Applied Biosystems, Foster City, CA). The matrix solution was saturated 3,5-dimethoxy-4-hydroxycinnamic acid in 50% acetonitrile with 0.1% formic acid for peptide analyses and sinapinic acid for protein analyses. The mass spectrometer was calibrated with insulin, cytochrome c, myoglobin, and BSA in linear mode or with trypsin autolysis peptides in reflectron mode.
LC/MS/MS
Samples (0.2 μl) were injected into a Micromass CapLC liquid chromatography system (Micromass, Manchester, UK) and pre-concentrated on a 300 μm × 5 mm PepMap C18 precolumn. The sample on the precolumn was washed for 3 min with 0.1% formic acid at a flow rate of 30 μL/min, eluted onto an analytical C18 column (150 mm × 17μm), and then analyzed with a solvent gradient from solution A (3% acetonitrile) to Solution B (95% acetonitrile) containing 0.1% formic acid over 50 min at a flow rate of 200 nL/min. The flow rate from pumps A and B was 5 μL/min and was reduced to 200 nL/min by stream splitting.
The LC eluant was introduced into the nanoflow source interfaced with a Micromass Q-TOF Micro mass spectrometer (Micromass, Manchester, UK). The source temperature was 80°C, and the cone gas flow was 50 L/hr. A voltage of 3.2 kV was applied to the nanoflow probe tip, and data were acquired in positive ion mode. The survey scans were integrated over one second, and the MS/MS scans were integrated over three seconds. Switching from survey scan to MS/MS scan mode was performed in a data-dependent manner. The maximum MS/MS scan to survey scan ratio was three. The collision energy was 28 eV. Data were processed using Masslynx version 3.5 software. Multi-point calibration was performed using selected fragment ions that resulted from the collision-induced decomposition of Glu-fibrino-peptide B. MS/MS spectra were processed by Masslynx software to produce a peak list file.
Signature-Discovery Algorithm
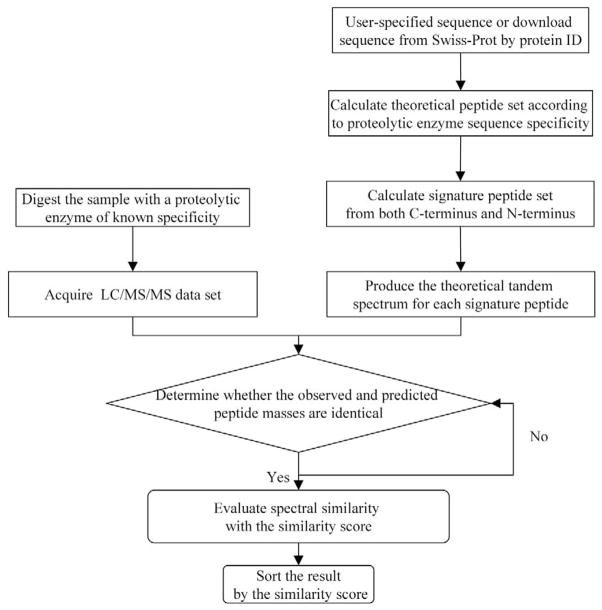
Figure 2 is the flow chart of the program to identify signature peptides derived from protein isoforms or variants. The theoretical set of peptides and their amino acid sequences are generated from the known sequence of the target protein and the sequence specificity of the protease used for digestion. A set of potential signature peptides and their sequences is then generated from the set of peptides produced from digestion of the target protein. The algorithm generates the signature peptide set by removing one amino acid sequentially from the N- or C-terminus of each expected peptide in the digest of the target protein. This potential signature peptide set will thus include peptides that arise from either the N- or C-terminus or from somewhere in the middle of the sequence. The algorithm would not detect products of unexpected splice variant mRNA species that contain some region of internal sequence not encoded by the cDNA species to which the peptide data is compared. If specific splice variants are known to exist, the peptide data set can be compared with the deduced amino acid sequence of more than one splice variant cDNA species.
Figure 2.
Flow chart of the Signature-Discovery Program.
Generating the potential signature peptide set from proteolytic peptides rather than from the entire amino acid sequence of the target protein dramatically reduces the size of the theoretical signature peptide set but does not sacrifice sequence coverage. If the target protein has 700 amino acid residues, for example, over 260,000 different potential signature peptides will be generated from the entire amino acid sequence using commercial software [23], but our algorithm generates only 820 candidate peptides. This reduces the complexity of the calculation by over 300-fold.
To include all potential signature peptides in the theoretical set, we allow up to five missed cleavages, and we use multiple proteases with different sequence specificity. The theoretical MS/MS spectrum of each candidate signature peptide is then generated from peptide CAD fragmentation rules. The solution containing the target protein and its variants is then digested with the protease selected for generation of the theoretical data set, and the resultant digest is analyzed by LC/MS/MS. Acquired and theoretical MS/MS spectra are then compared with each other in order to select signature peptide candidates in a two-step procedure. First, the molecular weights of peptides from the theoretical and actual data sets are compared. If peptides with identical molecular weights (within an error limit) are found in the two data sets, then the similarity of their MS/MS spectra is evaluated by an algorithm that generates a similarity score (eq 1 in the next section). Peptides with a high similarity score are identified as candidate signature peptides. Finally, all the selected peptides are sorted by the similarity score, and a higher score reflects a greater likelihood of correct identification.
Results and Discussion
Score Calculation
For a given molecular weight, there can be more than one candidate matching peptide, and each will have an amino acid sequence that differs from that of the others [40]. The accuracy of the similarity score is thus important in correctly identifying signature peptides. Generally, individual fragment ions of similar m/z value from the acquired and theoretical spectra are compared pair by pair. If the m/z difference between the two ions is less than the preset error tolerance, that fragment ion will be included in the similarity score calculation [41]. Peptide fragmentation information (first item in eq 1 below) is the most important component of the similarity score calculation. If the spectrum is noisy, fragmentation information alone is insufficient to identify the correct sequence, and intensity information (second item in eq 1 below) must also be considered in the similarity score calculation. In our similarity score calculation algorithm, the intensity information is included by counting the matched fragment ions in the 20 most intense spectral lines in the acquired spectrum.
| (1) |
Nmatch is the number of matched spectral lines in the theoretical and actual MS/MS data. Ntheoretical is the number of spectral lines in the theoretical MS/MS spectrum. Nmatch20 is the number of matched spectral lines in the 20 most intense spectral lines in the acquired MS/MS spectrum. The parameter k is a correction factor, and . It is used to correct for discrimination against smaller peptides when counting the matched fragment ions among the 20 most intense spectral lines.
To evaluate the similarity score calculation algorithm and to select an appropriate similarity score threshold, the standard proteins α-casein, cytochrome c, apomyoglobin, BSA, lysozyme and thyroglobulin were incubated with a mixture of two proteases (trypsin and Glu-C) at 37°C for 12 h. The digests were then analyzed by LC/MS/MS, and Masslynx 3.5 software was used to produce a peak list file for the MS/MS spectra. With a mixture of two proteases, the digest should contain three different kinds of peptides: (1) peptides generated by trypsin cleavage; (2) peptides generated by Glu-C cleavage; and (3) peptides in which one terminus is generated by trypsin cleavage and the other by Glu-C cleavage, which are designated as dual-enzyme cleavage peptides.
Because such dual-enzyme peptides contain N- and C-termini produced by different proteases, they represent surrogate signature peptides that can be used to optimize the experimental similarity score threshold. Table 1 lists the entire set of potential dual-enzyme peptides identified by our SD program and their similarity scores. Comparisons by direct inspection of the corresponding acquired MS/MS spectrum to the amino acid sequence deduced by our SD program resulted in selection of 0.2 as an appropriate similarity score threshold value. That value is used in subsequent calculations. Comparisons that yield a similarity score that is less than 0.2 are not considered acceptable matches, but, if desired, such spectra could be further evaluated by direct inspection.
Table 1.
Dual enzyme peptides identified by the Signature-Discovery program from digests of a mixture of model proteins
| Sequence | Expected Mass | Start AA | End AA | Calculated mass | Observed mass | Charge state | Item1a | Item2a | Similarity score |
|---|---|---|---|---|---|---|---|---|---|
| (E)WQQVLNVWGK(a) | 1257.6750 | 7 | 16 | 1257.6654 | 629.3327 | 2 | 0.50 | 0.70 | 0.60 |
| (E)REDLIAYLK(c) | 1120.6359 | 91 | 99 | 1120.6324 | 560.6162 | 2 | 0.43 | 0.75 | 0.59 |
| (E)NFVAFVDK(b) | 939.4942 | 573 | 580 | 939.4906 | 470.2483 | 2 | 0.40 | 0.70 | 0.55 |
| (R)LFTGHPETLE(a) | 1143.5686 | 32 | 41 | 1143.5632 | 572.2816 | 2 | 0.53 | 0.55 | 0.54 |
| (K)YKVPQLE(s) | 875.4827 | 119 | 125 | 876.4648 | 438.7324 | 2 | 0.81 | 0.25 | 0.53 |
| (K)EPMIGVNQE(s) | 1016.4735 | 148 | 156 | 1016.4514 | 508.7257 | 2 | 0.61 | 0.40 | 0.51 |
| (D)YGILQINSR(l) | 1063.5897 | 71 | 79 | 1063.5848 | 532.2924 | 2 | 0.46 | 0.55 | 0.51 |
| (D)LLIGSSQDDGLINR(t) | 1500.8026 | 2507 | 2520 | 1500.799 | 750.8995 | 2 | 0.39 | 0.60 | 0.50 |
| (K)VEADIAGHGQE(a) | 1125.5183 | 17 | 27 | 1125.504 | 563.252 | 2 | 0.42 | 0.55 | 0.49 |
| (E)YGFQNALIVR(b) | 1180.6480 | 424 | 433 | 1180.6474 | 590.8237 | 2 | 0.37 | 0.60 | 0.48 |
| (R)HPYFYAPE(b) | 1023.4583 | 169 | 176 | 1023.4616 | 512.2308 | 2 | 0.35 | 0.55 | 0.45 |
| (E)FISDAIIHVLHSK(a) | 1479.8326 | 106 | 118 | 1479.8269 | 493.9423 | 3 | 0.31 | 0.50 | 0.40 |
| (K)HQGLPQEVLNE(s) | 1263.6335 | 23 | 33 | 1263.5676 | 632.2838 | 2 | 0.35 | 0.45 | 0.40 |
| (E)DYLSLILNR(b) | 1106.6207 | 474 | 482 | 1106.6392 | 553.8196 | 2 | 0.31 | 0.45 | 0.38 |
| (R)KVPQVSTPTLVE(b) | 1297.7373 | 437 | 448 | 1297.7684 | 649.3842 | 2 | 0.33 | 0.40 | 0.37 |
| (E)LLYYANK(b) | 884.4884 | 177 | 183 | 884.4962 | 442.7481 | 2 | 0.33 | 0.40 | 0.36 |
| (D)SGIFQPMLQGR(t) | 1233.6415 | 419 | 429 | 1233.647 | 617.3235 | 2 | 0.33 | 0.25 | 0.29 |
| (R)FFVAPFPE(s) | 953.4785 | 38 | 45 | 953.4628 | 477.2314 | 2 | 0.22 | 0.30 | 0.26 |
| (K)EGIHAQQKEPMIGVNQE(s) | 1907.9297 | 140 | 156 | 1907.9008 | 636.6336 | 3 | 0.19 | 0.30 | 0.25 |
| (E)ADIAGHGQEVLIR(a) | 1378.7443 | 19 | 31 | 1378.8386 | 689.9193 | 2 | 0.21 | 0.20 | 0.21 |
| (E)LSVLLPNRQGLKK(t) | 1465.9215 | 2701 | 2713 | 1465.7136 | 733.3568 | 2 | 0.21 | 0.21 | 0.21 |
| (E)DLIAVLK(c) | 835.4925 | 93 | 99 | 835.4796 | 418.2398 | 2 | 0.13 | 0.30 | 0.21 |
| (E)DQSCPSERRR(t) | 1233.5753 | 361 | 370 | 1233.647 | 617.3235 | 2 | 0.21 | 0.10 | 0.15 |
| (K)ECCHGDLLECAD(b) | 1307.4707 | 267 | 278 | 1307.7456 | 654.3728 | 2 | 0.22 | 0.00 | 0.11 |
| (E)YAVSVLLR(b) | 920.5582 | 364 | 371 | 920.5558 | 460.7779 | 2 | 0.04 | 0.10 | 0.07 |
a: Apo-myoglobin, c: Cyto-chrome-C, b: Bove serum albumin, s: α-Casein, t: Thyroglobulin, l: Lysozyme.
Item 1 is the calculated value of first item in eq 1; Item 2 is the calculation value of first item in eq 1.
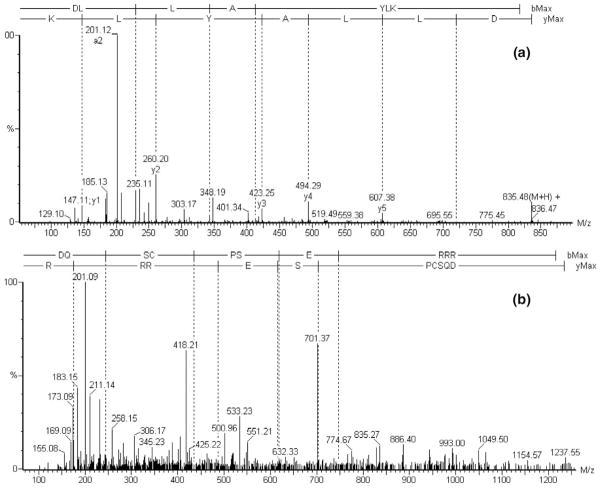
If the quality of MS/MS spectrum is poor (e.g., few fragment ions are recorded or the spectrum is noisy), intensity information (the second item in eq 1 above) plays a critical role in correct identification of dual-enzyme peptides. Figure 3a is the MS/MS spectrum of the identified dual-enzyme peptide (E)DLIAYLK from cytochrome c. Because too few fragment ions are recorded in the MS/MS spectrum, the calculated value of the first item in eq 1 (the contribution of its fragmentation information) is only 0.13, but the predicted fragment ions of this peptide (especially the y-ions) match most of the high intensity peaks in the observed spectrum. This causes the second item (eq 1, intensity information) to have a calculated value of 0.3. This peptide is thus identified as a dual-enzyme peptide with a similarity score of 0.21. Although the MS/MS spectrum (Figure 3b) of peptide (E)DQSCPSERRR from thyroglobulin has more fragment ions, most high intensity peaks do not match predicted fragment ions, and the similarity score in this case is lower when both the fragment and intensity data are considered together.
Figure 3.
The MS/MS spectrum of potential dual enzyme peptides: (a) MS/MS spectrum of peptide (E)DLIAYLK with a similarity score of 0.21; (b) MS/MS spectrum of peptide (E)DQSCPSERRR with a similarity score of 0.15.
Identification of Four Different Isoforms of Group VIA Phospholipase A2 expressed from its cDNA
The Group VIA Phospholipase A2 (iPLA2β) enzyme is a recently recognized member of the PLA2 family that has been proposed to participate in signaling, membrane remodeling, exocytosis, cell proliferation, apoptosis, and other events, and isoforms expressed in some cells are generated by proteolytic processing [3–10]. We have prepared recombinant constructs of iPLA2β that contain an N-terminal polyhistidine sequence (His-tag) or a C-terminal His-tag that will interact with immobilized metal affinity matrices, and these constracts have been expressed from their cDNAs in an Sf9 insect cell-baculovirus system.
The two constructs appeared to yield markedly different amounts of protein in this system after product purification by cobalt affinity column chromatography (Figure 4). The N-terminal half of the iPLA2β protein sequence contains seven ankyrin-like repetitive sequence stretches, and the C-terminal portion contains the catalytic site and some regulatory domains. We postulated that the low yield of the N-terminal His-tag iPLA2β from cobalt affinity columns might reflect N-terminal processing and loss of the His-tag, which would prevent interaction with the cobalt affinity column and cause loss of the product during purification. To evaluate this possibility, both SDS-PAGE bands illustrated in the rightmost lane of Figure 4 were excised and digested with trypsin.
Figure 4.
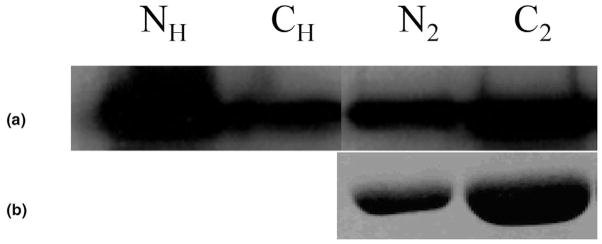
Immunoaffinity (a) and Coomassie blue stain (b) of SDS PAGE analysis of eluants from cobalt affinity columns. N: N-terminal His-tag iPLA2β; C: C-terminal His-tag iPLA2β. Subscript H: homogenate, 2: the second eluant.
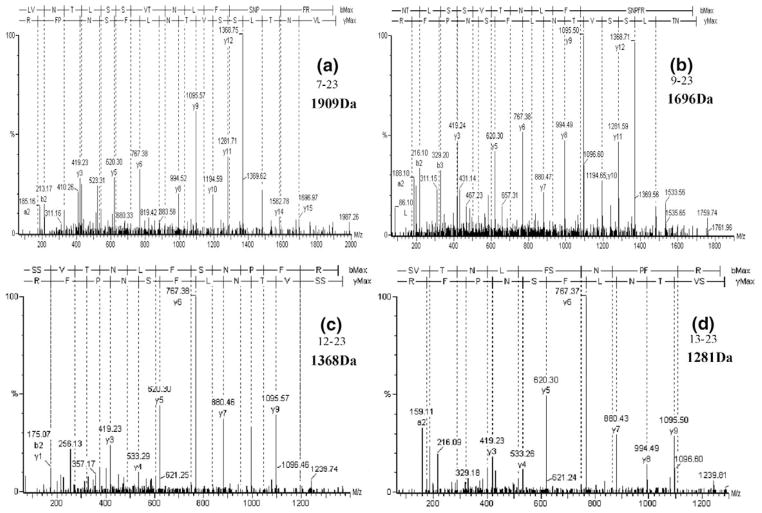
The digests were analyzed by MALDI/TOF/MS to acquire the peptide mass map and by LC/MS/MS. The first 30 N-terminal amino acids of iPLA2β are 1MQFFGRLVNT 11LSSVTNLFSN 21PFRAKEVSLA. The N-terminal tryptic peptide of the full-length iPLA2β isoform is 1MQFFGR6, but this peptide is N-acetylated and generates only a singly charged ion that does not trigger switching from survey MS mode to MS/MS mode on the Q-TOF [24]. The second tryptic peptide in full-length iPLA2β is 7L-R23. Peptides with residue 23 as the C-terminus and residues 8 or higher as the N-terminus represent signature peptides that reflect N-terminal proteolytic processing. Analyses of the MS/MS data of the tryptic peptide mixture from C-terminal His-tag iPLA2β by the SD program results in identification of three different signature peptides (Table 2) that represent different truncation variants (9N-P752, 12S-P752, and 13S-P752). Figure 5 illustrates the MS/MS spectra of the corresponding signature peptides (9N-R23, 12S-R23, and 13S-R23) and that of the tryptic peptide 7L-R23 of full-length iPLA2β. The high correspondence between the fragment ions in each spectrum and the predicted amino acid sequence demonstrates accurate identification of these signature peptides. All three signature peptides are also observed in the MALDI/TOF mass spectrum of the tryptic digest of C-terminal His-tagged iPLA2β (Figure 6a). The signature peptide 12SSVTNLFSNPFR23 exhibits the most intense signal, and the signal of the tryptic peptide 7LVNTLSSVTNLFSNPFR23 of the full-length iPLA2β is only 10% of the sum of the signals for the three other signature peptides.
Table 2.
Signature peptides identified by the Signature-Discovery program from a trypsin digest of iPLA2β
| Sequence | Expected mass | StartAA | EndAA | Calculated mass | Observed mass | Charge state | Similarity score |
|---|---|---|---|---|---|---|---|
| SSVTNLFSNPFR | 1368.691 | 12- | 23 | 1368.629 | 684.8147 | 2 | 0.58 |
| SVTNLFSNPFR | 1281.659 | 13- | 23 | 1281.622 | 641.3112 | 2 | 0.54 |
| NTLSSVTNLFSNPFR | 1696.866 | 9- | 23 | 1696.814 | 848.9069 | 2 | 0.50 |
Figure 5.
Tandem mass spectra of different signature peptides identified in tryptic digest of iPLA2β with a C-terminal His-tag: (a) the tandem mass spectrum of the expected peptide from the full length isoform (7L-R23); (b), (c), and (d), the tandem mass spectra of signature peptides (9N-R23, 12S-R23, and 13S-R23) that represent truncated isoforms.
Figure 6.
MALDI/TOF mass spectrum of the peptide mixture from in-gel digestion of the His-tagged iPLA2β bands in Figure 3, (a) C-terminal His-tag iPLA2β; (b) N-terminal His-tag iPLA2β.
Because much of the amino acid sequence of the four peptides is identical, the relative abundances of the different variants is likely to be approximately reflected by the relative signal intensities of their signature peptides. The major variant expressed in the baculovirus Sf9 cell system thus appears to be (9N-P752), and full-length iPLA2β appears to be a minor component of the iPLA2β isoform mixture. This is consistent with Edman sequencing of recombinant iPLA2β expressed in Sf9 cells [3]. Figure 6b is the MALDI/TOF mass spectrum of the tryptic digest of N-terminal His-tag iPLA2β and illustrates that the only N-terminal peptide from full-length iPLA2β observed in the tryptic digest is 7LVNTLSSVTNLFSNPFR23. All three of the iPLA2β N-terminal truncation variants lack the His-tag and thus fail to interact with the cobalt affinity column, which results in their loss during affinity purification.
Identification of Potential Cleavage Sites of the C-Terminal Fragments from an Amyloid Precursor Protein (APP) Construct
The SD program was also applied to the more complex biological problem of identifying the cleavage sites of Amyloid Precursor Protein (APP) involved in the pathogenesis of Alzheimer’s Disease [1]. These sites are reflected by the N-terminal residue of the six myc-tagged C-terminal APP fragment that results from the cleavages. This fragment is also designated either as the the APP intracellular domain (AICD) or as the carboxy-terminal fragment generated by γ secretase cleavage (CTFγ). The sequence of the six myc-tagged AICD peptide of APP is 1LVMLKKKQYT 11SIHHGVVEVD 21AAVTPEERHL 31SKMQQNGYEN 41PTYKFFEQMH 51RFKAMEQKLI 61SEEDLNEMEQ 71KLISEEDLNE 81MEQKLISEED 91LNEMEQKLIS 101EEDLNEMEQK 111LISEEDLNEM 121ESLGDLTMEQ 131KLISEEDLNS 141RPLEPLEL, and its MW about 17 kDa.
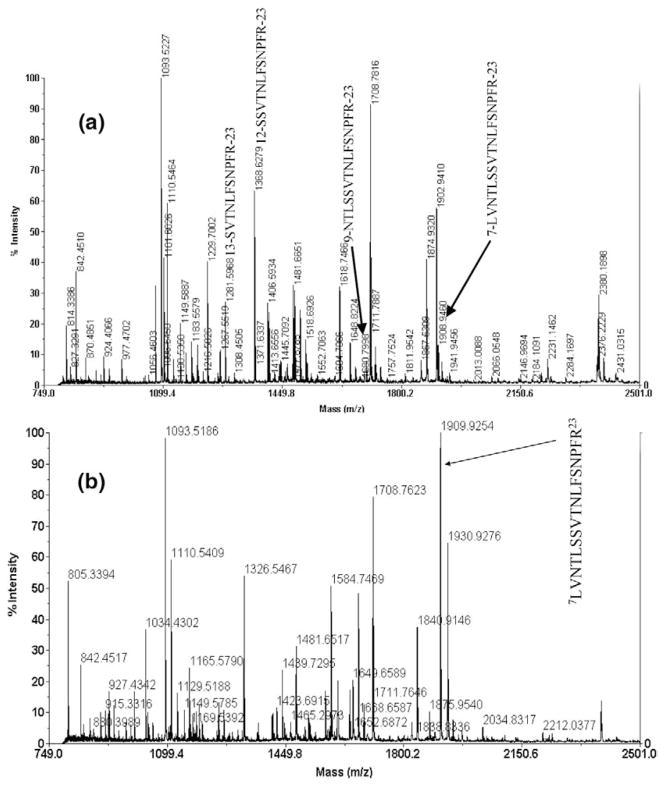
The sequence that corresponds to the six-myc tag is 54M-L148. This tag is added to the APP construct to facilitate immunoprecipitation from homogenates after expression in transfected cells with antibodies directed against the tag sequence. APP can undergo cleavage at different sites within cells to produce different C-terminal fragments. Because APP immunoprecipitates (IP) obtained with agarose beads coupled to antibodies directed against its myc-tag epitope contain many components, it is impossible to identify the APP cleavage sites from the MALDI/TOF mass spectrum of the IP (Figure 7). We therefore digested the eluate from the IP beads with trypsin and Glu-C separately. The digests were then analyzed by LC/MS/MS, and the data were processed by our SD program.
Figure 7.
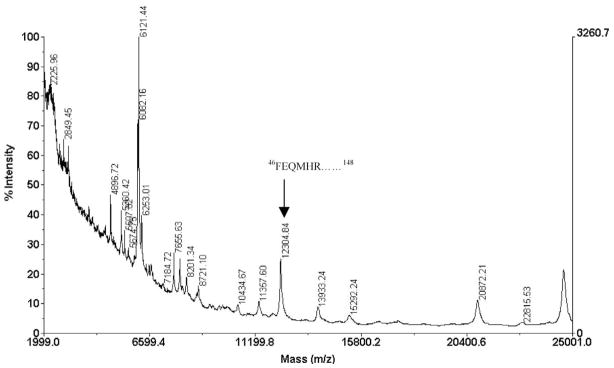
MALDI/TOF mass spectrum of the eluants from immunoprecipitates of C-terminal six myc-tagged APP cleavage products. The arrow identifies the peak that corresponds to the truncated variant of the C-terminal fragment starting with amino acid 46F as the N-terminus.
The signature peptides identified from the tryptic digest are listed in Table 3. The peptide 46FEQMHR51 arises from a nontryptic cleavage, and the similarity score of its MS/MS spectrum (Figure 8) is 0.34. Most of the expected theoretical fragment ion m/z values match spectral lines in Figure 8 within ±0.1 m/z units (Table 4). Observed high abundance ions match several expected fragment ions in the theoretical y-series (y1, 175.12; y2, 312.21; y3, 443.31, y4, 571.33, and y5, 700.42) and in the z-series (z2, 295.18; z3, 426.25; z4, 554.30; and z5, 683.40). Several predicted ions in the a-series and in the b-series of fragment ions also match intense peaks in Figure 8 (Table 4). The MS/MS spectrum in Figure 8 thus supports accurate identification of a nontryptic cleavage product by the SD algorithm.
Table 3.
Signature peptide identified from the tryptic digest of APP six myc-tagged C-terminal cleavage fragment
| Sequence | Expected mass | StartAA | EndAA | Calculated mass | Observed mass | Charge state | Similarity score |
|---|---|---|---|---|---|---|---|
| FEQMHR | 847.3889 | 46- | 51 | 847.4916 | 424.2458 | 2 | 0.34 |
| ESLGDLTMEQK | 1250.593 | 121- | 131 | 1250.725 | 417.575 | 3 | 0.03 |
Figure 8.

MS/MS spectrum of the signature peptide 46FEQMHR51 from a tryptic digest of APP six myc-tagged C-terminal cleavage products.
Table 4.
The theoretical fragment masses of signature peptide 46FEQMHR51 and the m/z tolerance of experimental fragment masses in Figure 8
| a ions | Theoretical m/z | 120.08 | 249.12 | 377.18 | 508.22 | 645.28 | 801.38 |
| m/z tolerance | −0.00 | −0.01 | --- | 0.07 | −0.01 | --- | |
| b ions | Theoretical m/z | 148.08 | 277.12 | 405.18 | 536.22 | 673.28 | 829.38 |
| m/z tolerance | −0.10 | −0.05 | −0.10 | −0.09 | --- | --- | |
| AA | Phe | Glu | Gln | Met | His | Arg | |
| y ions | Theoretical m/z | 847.39 | 700.32 | 571.28 | 443.22 | 312.18 | 175.12 |
| m/z tolerance | --- | −0.10 | −0.06 | −0.09 | −0.03 | −0.02 | |
| z ions | Theoretical M/z | 830.36 | 683.29 | 554.25 | 426.19 | 295.15 | 158.09 |
| m/z tolerance | - | −0.11 | −0.05 | −0.07 | −0.03 | −0.08 |
Re-examination of the MALDI/TOF mass spectrum of the original immunoprecipitate in Figure 7 reveals a peak at m/z 12,304.84 closely matches the predicted m/z value (12,304.73) of the C-terminal cleavage fragment 46F-L148, which is the expected parent polypeptide from which the 46F-R51 fragment arises upon digestion with trypsin. All of this information supports the identification of a potential C-terminal fragment of the APP construct with a C-terminal six-myc tag that is generated by cleavage of the 45F-F46 peptide bond, and this is a cleavage site that has not been recognized previously.
Results from digestion of the immunoprecipitate with the alternate protease Glu-C are summarized in Table 5. The first listed peptide is a potential signature peptide with a similarity score of 0.30. Its tandem mass spectrum (Figure 9) contains high intensity peaks that match predicted fragment ions of a candidate signature peptide. In the signature peptide sequence 45FEQMHR51 identified in Table 3, one glutamic acid residue is located two amino acid residues away from the N-terminal phenylalanine residue. The N-terminal signature peptide produced by GluC digestion would be the dipeptide FE, which is too small to be captured on the C18 LC column and then detected by the mass spectrometer. That is the reason why the cleavage site 45F-F46 is not identified in analyses of the Glu-C digest. This illustrates why we recommend digesting with more than one protease and then analyzing the MS/MS data of different digests with the SD program in order to avoid overlooking potential signature peptides.
Table 5.
Signature peptide identified from the Glu-C digest of APP six myc-tagged C-terminal cleavage fragment
| Sequence | Expected mass | StartAA | EndAA | Calculated mass | Observed mass | Charge state | Similarity score |
|---|---|---|---|---|---|---|---|
| YENPTYKFFE | 1337.605 | 38 | 47 | 1337.683 | 669.3414 | 2 | 0.30 |
| MQQNGYENPTYKFFE | 1895.827 | 33 | 47 | 1895.947 | 948.4734 | 2 | 0.16 |
| AMEQKLISEE | 1177.578 | 54 | 63 | 1177.753 | 589.3766 | 2 | 0.10 |
| LKKKQYTSIHHGVVEVDAAVTPE | 2549.374 | 4 | 26 | 2549.364 | 850.4546 | 3 | 0.03 |
| QMHRFKAME | 1177.561 | 48 | 56 | 1177.753 | 589.3766 | 2 | 0.02 |
| LKKKQYTSIHHGVVEVDAAVTPE | 2549.374 | 4 | 26 | 2549.375 | 1275.188 | 2 | 0.01 |
Figure 9.
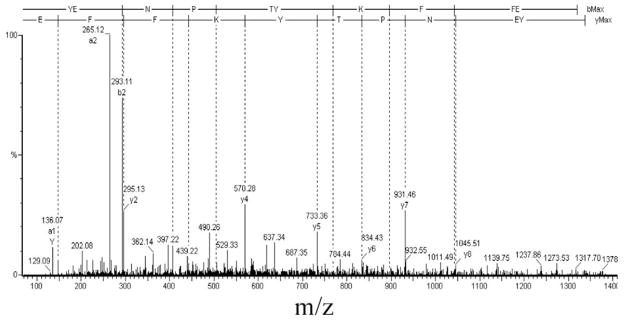
MS/MS spectrum of the signature peptide 38YENPTYKFFE47 from a Glu-C proteolytic digest of APP six myc-tagged C-terminal cleavage products.
Limitations of the Signature Peptide Discovery Algorithm in Its Current Form
A caveat is that occasional nonspecific cleavage at unexpected sites by trypsin or other proteases is a limitation of any approach that relies on the sequence specificity of a given protease, and many searching algorithms in proteomics do so [23, 24]. Other such algorithms and ours can be confounded by such non-specific cleavages. The effect is that some apparent “signature peptides” do not truly reflect in vivo processing but are artifacts of nonspecific cleavages. The observed “signature peptide set” must thus be assumed to include some false positives, and a second piece of information consistent with the proposed site of processing is required as confirmation.
In the case of the APP peptide 46FEQMHR51, for example, that reflects cleavage of the 45F-F46 peptide bond, the fact that this is not a nonspecific tryptic cleavage is indicated by finding that the original immunoprecipitate that had not yet been digested with trypsin contained the parent polypeptide 45F-L148 from which the tryptic product 46FEQMHR51 was subsequently produced. Similarly, the fact that the peptide 12SSVTNLFSNPFR23 observed in purified iPLA2β digests does not reflect nonspecific tryptic cleavage is indicated by the fact that Edman sequencing of the undigested preparation identifies 12S as the N-terminal residue.
It is also important when comparing actual and theoretical mass spectra that true matches should be distinguished from random hits [25, 42], and some software programs, e.g., Proteometrix Sonar (Genomics Solutions) or Mascot (Matrix Science), use other matching algorithms. In our particular application, the problem of random hits is considerably reduced by the fact that our peptide tandem mass spectra are not searched against entire protein data bases but rather against the sequence of a specific protein, and this greatly decreases the family of random hit candidates that have the same parent ion m/z value (within a specified tolerance) and somewhat similar MS/MS spectra but different amino acid sequences than the actual peptide from which the experimental MS/MS spectrum was produced.
Our algorithm does consider peak intensity information in addition to m/z value and also includes a correction factor designed to minimize discrimination against smaller peptides, but it still represents an enhanced Signal Dot Product (SDP) approach. Both Sequest and Sonar MS/MS also use SDP as the basic scoring algorithm [43]. The Sonar MS/MS algorithm includes a parameter in comparing spectra designated the “expectation value” that is designed to enhance discrimination between random hits and true matches, and it is based on the probability distribution of stochastic SDP scores of peptide sequences [42]. Because our application focuses on the sequence of a single protein rather than an entire protein database, there is an insufficient number of stochastic scores to calculate this distribution function.
Conclusions
In the course of studying in vivo processing of the central nervous system proteins Group VIA Phospholipase A2 (iPLA2β) and Amyloid Precursor Protein (APP), and we have developed an automated method to identify protein isoforms that arise from proteolytic processing. Algorithms for generating sets of theoretical signature peptides and for calculating similarity scores to compare actual and theoretical mass spectra in our Signature-Discovery (SD) program have been optimized, and appropriate parameters have been selected from experiments with model protein mixtures. Using our optimized parameters, four different isoforms of iPLA2β produced by proteolytic processing have been identified among products derived from expression of iPLA2β cDNA in cells. We have also used the SD program to identify two variants of the C-terminal cleavage fragment of an APP construct with a C-terminal six-myc tag that represent previously unrecognized cleavage sites for APP processing. These results with iPLA2β and APP and illustrate the utlility of our approach in identifying protein isoforms that arise from proteolytic processing.
Acknowledgments
This work was supported by grants R37-DK34388, P01-HL57278, P41-RR00954, P60-DK20579, and P30-DK56341.
References
- 1.Hardy J, Selkoe DJ. The Amyloid Hypothesis Alzheimer’s Disease: Progress and Problems on the Road to Therapuetics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Kopan R, Goate A. A Common Enzyme Connects Notch Signaling and Alzheimer’s Disease. Genes Dev. 2000;14:2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 3.Ramanadham S, Song H, Bao S, Hsu FF, Zhang S, Ma Z, Jin C, Turk J. Islet Complex Lipids: Involvement in Group VIA Calcium-Independent Phospholipase A2-Mediated Effects in β-Cells. Diabetes. 2004;53(Suppl 1):S179–S185. doi: 10.2337/diabetes.53.2007.s179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Z, Wang X, Nowatzke W, Ramanadham S, Turk J. Human Pancreatic Islets Express mRNA Species Encoding Two Distinct Catalytically Active Isoforms of Group VI Phospholipase A2 (iPLA2) that Arise from an Exon-Skipping Mechanism of Alternative Splicing of the Ttranscript from the iPLA2 Gene on Chromosome 22q13.1. J Biol Chem. 1999;274:9607–9616. doi: 10.1074/jbc.274.14.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramanadham S, Hsu FF, Bohrer A, Ma Z, Turk J. Studies of the Role of Group VI Phospholipase A2 (iPLA2) in Fatty Acid Incorporation, Phospholipid Remodeling, Lysophosphatidylcholine Generation, and Secretagogue-Induced Arachidonic Acid Release in Pancreatic Islets and Insulinoma Cells. J Biol Chem. 1999;274:13915–13927. doi: 10.1074/jbc.274.20.13915. [DOI] [PubMed] [Google Scholar]
- 6.Ma Z, Ramanadham S, Wohltmann M, Bohrer A, Hsu FF, Turk J. Studies of Insulin Secretory Responses and of Arachidonic Acid Incorporation into Phospholipids of Stably Transfected Insulinoma Cells that Overexpress Group VIA Phospholipase A2 (iPLA2β) Indicate a Signaling Rather than a Housekeeping Role for iPLA2β. J Biol Chem. 2001;276:13198–13208. doi: 10.1074/jbc.M010423200. [DOI] [PubMed] [Google Scholar]
- 7.Ma Z, Bohrer A, Wohltmann M, Ramanadham S, Hsu FF, Turk J. Studies of Phospholipid Metabolism, Proliferation, and Secretion of Stably Transfected Insulinoma Cells that Overexpress Group VIA Phospholipase A2. Lipids. 2001;36:689–700. doi: 10.1007/s11745-001-0774-9. [DOI] [PubMed] [Google Scholar]
- 8.(a) Ma Z, Turk J. Molecular Biology of the Group VIA Ca2+-Independent Phospholipase. Prog Nucl Acid Res Molec Biol. 2001;67:1–33. doi: 10.1016/s0079-6603(01)67023-5. [DOI] [PubMed] [Google Scholar]; (b) Moldave K, editor. Academic Press: San Diego. [Google Scholar]
- 9.Ramanadham S, Song H, Hsu FF, Zhang S, Crankshaw M, Grant G, Newgard C, Turk J. Pancreatic Islets and Insulinoma Cells Express a Novel Isoform of Group VIA phospholipase A2 (iPLA2β) that Participates in Glucose-Stimulated Insulin Secretion and is not Produced by Alternate Splicing of the iPLA2β Transcript. Biochemistry. 2003;42:13929–13940. doi: 10.1021/bi034843p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramanadham S, Hsu FF, Zhang S, Jin C, Bohrer A, Ma Z, Turk J. Involvement of the Group VIA Phospholipase A2 (iPLA2β) in Endoplasmic Reticulum Stress-Induced Apoptosis in Insulinoma Cells. Biochemistry. 2004;43:918–930. doi: 10.1021/bi035536m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey A, Mann M. Proteomics to Study Genes and Genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 12.Wickner S, Maurizi MR, Gottesman S. Post-Translational Quality CcontrolFolding, Refolding, and Degrading Proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 13.Weihofen A, Martoglio B. Intramembrane-Cleaving Proteases: Controlled Liberation of Proteins and Bioactive Peptides. Trends Cell Biol. 2003;13:71–78. doi: 10.1016/s0962-8924(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 14.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein Function in the Post-Genomic Era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 15.Wang R, Sweeney D, Gandy SE, Sissodia SS. The Profile of Soluble Amyloid β Protein in Cultured Cell Media. Detection and Quantification of Amyloid β Protein and Variants by Immunoprecipitation Mass Spectrometry. J Biol Chem. 1996;271:31894–31902. doi: 10.1074/jbc.271.50.31894. [DOI] [PubMed] [Google Scholar]
- 16.Mann M, Hojrup P, Roepstorff P. Use of Mass-Spectrometric Molecular Weight Information to Identify Proteins in Sequence Databases. Biol Mass Spectrom. 1993;22:338–345. doi: 10.1002/bms.1200220605. [DOI] [PubMed] [Google Scholar]
- 17.Yates JR, Speicher S, Griffin PR, Hunkapiller T. Peptide Mass Maps. A Highly Informative Approach to Protein Identification. Anal Biochem. 1993;214:397–408. doi: 10.1006/abio.1993.1514. [DOI] [PubMed] [Google Scholar]
- 18.Mortz E, Vorm O, Mann M, Roepstorff P. Identification of Proteins in Polyacrylamide Gels by Mass-Spectrometric Peptide Mapping with Database Searching. Biol Mass Spectrom. 1994;23:249–261. doi: 10.1002/bms.1200230503. [DOI] [PubMed] [Google Scholar]
- 19.Mann M. Role of Mass Accuracy in the Identification of Proteins by their Mass-Spectrometric Peptide Maps. J Protein Chem. 1994;13:506–507. [Google Scholar]
- 20.Fenyo D, Qin J, Chait BT. Protein Identification Using Mass Spectrometric Information. Electrophoresis. 1998;19:998–1005. doi: 10.1002/elps.1150190615. [DOI] [PubMed] [Google Scholar]
- 21.Lahm HW, Langen H. Mass Spectrometry: A Tool for the Identification of Proteins Separated by Gels Electrophoresis. Electrophoresis. 2000;21:2105–2114. doi: 10.1002/1522-2683(20000601)21:11<2105::AID-ELPS2105>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Wagner K, Miliotis T, Marko-Varga G, Bischoff R, Unger KK. An Automated On-Line Multidimensional HPLC System for Protein and Peptide Mapping with Integrated Sample Preparation. Anal Chem. 2002;74:809–820. doi: 10.1021/ac010627f. [DOI] [PubMed] [Google Scholar]
- 23.Eng JK, Mccormack AL, Yates JR. An Approach to Correlate Peptide MS/MS Data with Amino Acid Squences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 24.Mann M, Wilm M. Error Tolerant Identification of Peptides in Sequence Databases by Peptide Sequence Tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 25.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-Based Protein Identification by Searching Sequence Databases Using Mass Spectrometry Data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-de-Cossio J, Gonzalez J, Betancourt L, Besada V, Padron G, Shimonishi Y, Takao T. Automated Interpretation of High-Energy Collision-Induced Dissociation Mass Spectra of Singly Protonated Peptides by SeqMS Software for de Novo Sequencing. Rapid Commun Mass Spectrom. 1998;12:1867–1878. doi: 10.1002/(SICI)1097-0231(19981215)12:23<1867::AID-RCM407>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 27.Horn DM, Zubarev RA, McLafferty FW. Automated de Novo Sequencing of Proteins by Tandem High-Resolution Mass Spectrometry. Proc Natl Acad Sci US A. 2000;97:10313–10317. doi: 10.1073/pnas.97.19.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor JA, Johnson RS. Implementation and Uses of Automated de Novo Peptide Sequencing by Tandem Mass Spectrometry. Anal Chem. 2001;73:2594–2604. doi: 10.1021/ac001196o. [DOI] [PubMed] [Google Scholar]
- 29.Wilkins MR, Gasteiger E, Gooley AA, Herbert BR, Molloy MP, Binz PA, Ou K, Sanchez JC, Bairoch A, Williams KL, Hochstrasser DF. High-Throughput Mass Spectrometric Discovery of Protein Post-Translational Modifications. J Mol Biol. 1999;289(3):645–657. doi: 10.1006/jmbi.1999.2794. [DOI] [PubMed] [Google Scholar]
- 30.Gatlin CL, Eng JK, Cross ST, Detter JC, Yates JR. Automated Identification of Amino Acid Sequence Variations in Proteins by HPLC/Microspray Tandem Mass Spectrometry. Anal Chem. 2000;72:757–763. doi: 10.1021/ac991025n. [DOI] [PubMed] [Google Scholar]
- 31.Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking Genome and Proteome by Mass Spectrometry: Large-Scale Identification of Yeast Proteins from Two-Dimensional Gels. Proc Natl Acad Sci US A. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kris G, Marc G, Lennart M, Jozef VD, An S, Grégoire RT, Joël V. Exploring Proteomes Analyzing Protein Processing by Mass Spectrometric Identification of Sorted N-Terminal Peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins CM, Wolf MJ, Mancuso DJ, Gross RW. Identification of the Calmodulin-Binding Domain of Recombinant Calcium-Independent Phospholipase A2β. Implications for Structure and Function. J Biol Chem. 2001;276:7129–7135. doi: 10.1074/jbc.M010439200. [DOI] [PubMed] [Google Scholar]
- 34.Nowatzke W, Ramanadham S, Hsu FF, Ma Z, Bohrer A, Turk J. Mass Spectrometric Evidence that Agents which Cause loss of Ca2+ from Intracellular Compartments Induce Hydrolysis of Arachidonic Acid from Pancreatic Islet Membrane Phospholipids by a Mechanism that Does Not Require a Rise in Cytosolic Ca2+ Concentration. Endocrinology. 1998;139:4073–4085. doi: 10.1210/endo.139.10.6225. [DOI] [PubMed] [Google Scholar]
- 35.Wolf MJ, Gross RW. Expression, Purification, and Kinetic Characterization of a Recombinant 80 kDa Intracellular Calcium-Independent Phospholipase A2. J Biol Chem. 1996;271:30879–30885. doi: 10.1074/jbc.271.48.30879. [DOI] [PubMed] [Google Scholar]
- 36.O’Reilly DR, Miller LK, Luckow VA. Baculovirus Expression Vector: A Laboratory Manual. Freeman and Co; New York: 1992. [Google Scholar]
- 37.Ma Z, Ramanadham S, Kempe K, Chi XS, Ladenson J, Turk J. Pancreatic Islets Express a Ca2+-Independent Phospholipase A2 Enzyme that Contains a Repeated Structural Motif Homologous to the Integral Membrane Protein Binding Domain of Ankyrin. J Biol Chem. 1997;272:11118–11127. [PubMed] [Google Scholar]
- 38.Bao S, Jin C, Zhang S, Turk J, Ma Z, Ramanadham S. The β-Cell Calcium-Independent Group VIA phospholipase A2 (iPLA2β). Tracking iPLA2β Movements in Response to Stimulation with Insulin Secretagogues in INS-1 Cells. Diabetes. 2004;53:S186–S189. doi: 10.2337/diabetes.53.2007.s186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Bunkan AL, Hecimovic S, Walker E, Goate A. Conserved “PAL” Sequence in Presenilins is Essential for γ-Secretase Activity but not Required for Formation or Stabilization of γ-Secretase Complexes. Neurobiol Dis. 2004;15:654–666. doi: 10.1016/j.nbd.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Goodlett DR, Bruce JE, Anderson GA, Rist B, Pasa-Tolic L, Fiehn O, Smith RD, Aebersold R. Protein Identification with a Single Accurate Mass of a Cysteine-Containing Peptide and Constrained Database Searching. Anal Chem. 2000;72:1112–1118. doi: 10.1021/ac9913210. [DOI] [PubMed] [Google Scholar]
- 41.Pevzner P, Dancidk V, Tang C. Mutation-Tolerant Protein Identification by Mass Spectrometry. J Comput Biol. 2000;7:777–787. doi: 10.1089/10665270050514927. [DOI] [PubMed] [Google Scholar]
- 42.Fenyo D, Beavis RC. A Method for Assessing the Statistical Significance of Mass Spectrometry-Based Protein Identifications Using General Scoring Schemes. Anal Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 43.Yan F, Qiang Y, Ruixiang S, Ruixiang S, Dequan L, Rong Z, Charles XL, Wen G. Exploring the Kernel Trick to Correlate Fragment Ions for Peptide Identification via Tandem Mass Spectrometry. Bioinformatics. 2004;20:1948–1954. doi: 10.1093/bioinformatics/bth186. [DOI] [PubMed] [Google Scholar]