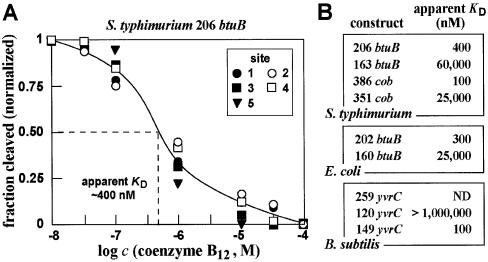
Figure 4.
Measurements of binding affinities of various coenzyme B12 aptamers. (A) Representative plot of the ligand concentration-dependent modulation of spontaneous RNA cleavage using the 206 btuB fragment from S.typhimurium. The fraction of RNA cleaved at sites 1–5 (Fig. 3C) was normalized against the fraction cleaved at each site when incubated in the absence of coenzyme B12. Site 5 is not present in the preceding image, but is present in the larger RNA construct used for this KD analysis (data not shown). (B) Comparison of the apparent KD values for various coenzyme B12 aptamer constructs from several bacteria. Values were derived for each construct using the approach described in (A).