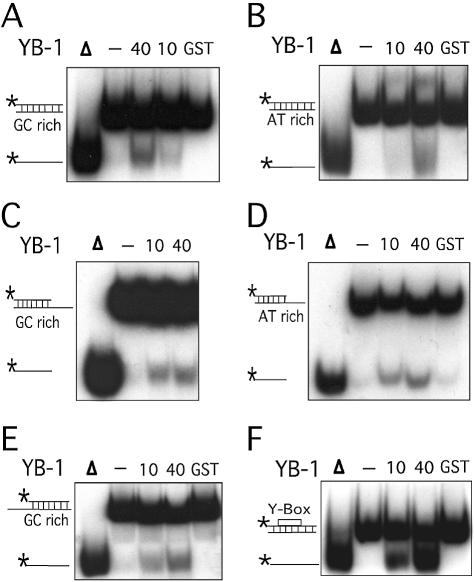
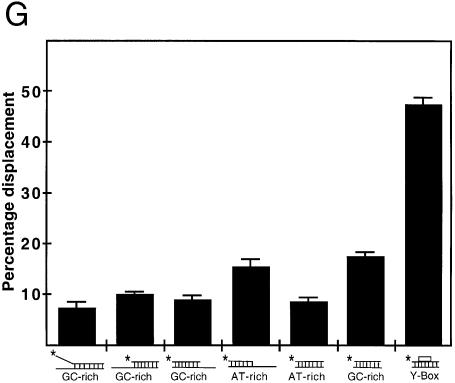
Figure 3.
Strand separation activity of YB-1 on different duplex structures. (A–F) The indicated amounts (in ng) of purified human YB-1 proteins were incubated with the indicated radioactive DNA substrates under standard conditions for helicase activity (see Materials and Methods) for 30 min at 37°C. Again, 40 ng of GST proteins were used as negative controls when indicated. Reactions were stopped in the appropriate dye buffer and the DNA products were run on a 12% native polyacrylamide gel. The double-stranded and single-stranded DNA structures with their sequence characteristics are depicted on the left of each autoradiogram. The 5′-labeled strand of the duplex is represented by an asterisk. (A) and (B) represent reactions with blunt-ended DNA. (C) and (D) represent reactions with a structure having a 3′ recessed end. (E) represents reactions with a structure having a 5′ recessed end. In (F), the Y-box sequence is an inverted CCAAT sequence. All DNA substrates contain 22 paired bases. (G) Histogram representation of the YB-1 strand separation reactions performed in (A–F). The strand displacement data were obtained from experiments done in duplicate with 40 ng of the indicated purified proteins. The percentage strand displacement is calculated by using the formula: c.p.m. of displaced strand/(c.p.m. of displaced strand + c.p.m. of double-stranded DNA) × 100. The c.p.m. was calculated for each band of the gel as indicated in Materials and Methods.