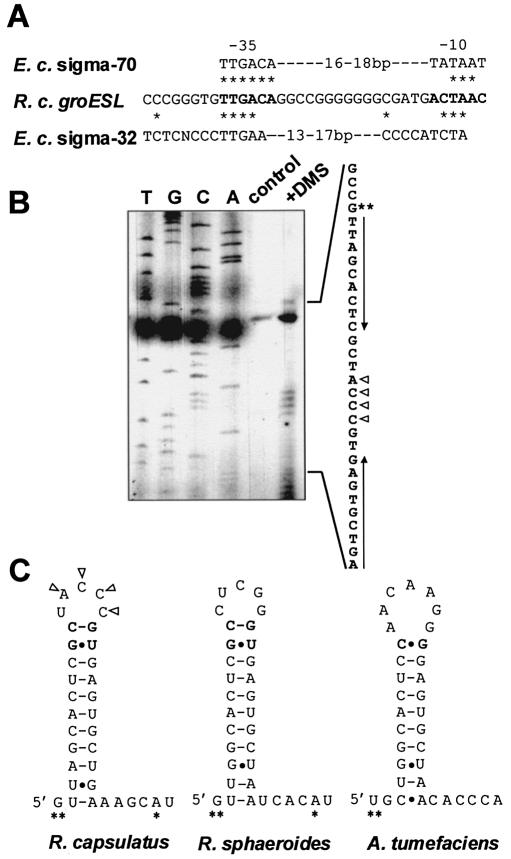
Figure 2.
Predicted promoter region of the R.capsulatus groESL operon and RNA secondary structure of CIRCE. (A) Homology of the putative groESL promoter sequence of R.capsulatus (R. c. groESL) to the consensus sequences of the E.coli vegetative sigma factor promoter (E. c. sigma-70) and the E.coli heat shock sigma factor promoter (E. c. sigma-32) (42). Identical residues are marked by asterisks. (B) Primer extension analysis after in vivo methylation was performed by adding DMS to a rapidly growing culture before RNA isolation. Methylated nucleotides were mapped by primer extension analysis (lane: +DMS) using the oligonucleotide PR-Cpn244- hybridizing to the groESL transcript ∼60–80 nt downstream of the major 5′-end. The modification blocks the primer extension reaction at the nucleotide preceding the modified residue. The control lane shows the extension products of unmodified RNA. Sequencing reactions obtained with the same oligonucleotide were run on a sequencing gel side by side with the products of the primer extension reaction and are labelled with T, G, C and A. Triangles indicate modified nucleotides. The major 5′-end is indicated by two asterisks and the palindromic CIRCE sequence is marked by arrows. (C) RNA secondary structure of the CIRCE region in the groESL 5′-untranslated region of three α proteobacteria. Modified nucleotides detected by in vivo methylation are indicated by triangles. Nucleotides of the variable region of CIRCE forming additional base pairs of the stem are in bold. 5′-ends are marked by asterisks, Watson–Crick base pairs are indicated by dashes and non-Watson–Crick base pairs by dots.