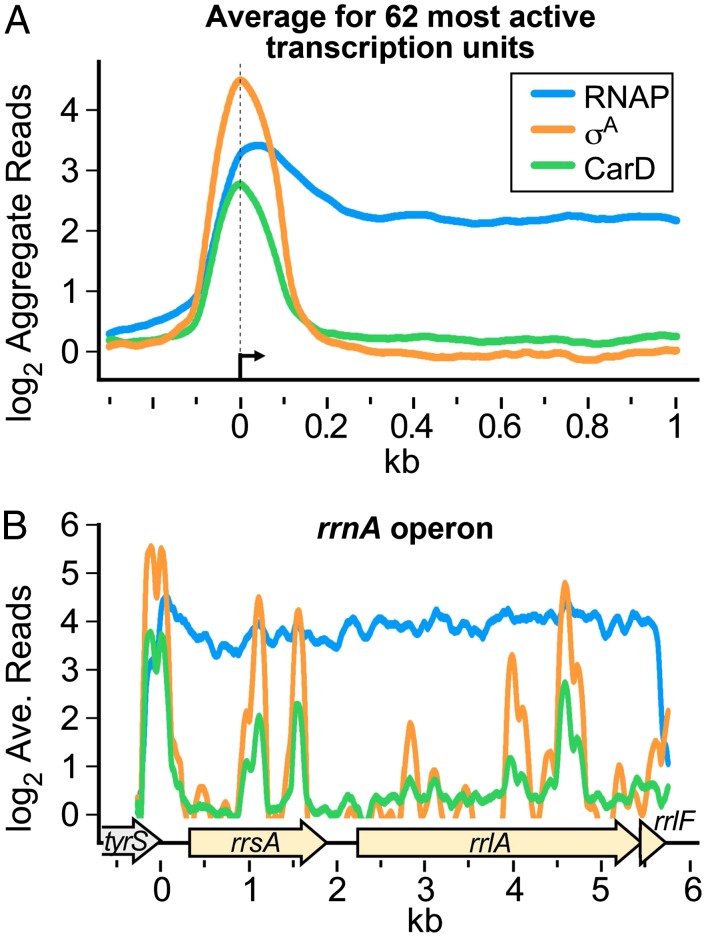
Fig. 1.
Normalized log2 of ChIP-seq reads from M. smegmatis DNA coimmunoprecipitated with RNAP β, σA, or CarD-HA. (A) Aggregate profiles averaged over 62 highly active transcription units. Protein–DNA complexes containing CarD-HA, RNAP β, and RNAP σ were immunoprecipitated from M. smegmatis lysates. The coprecipitated DNA was sequenced, and the number of sequence reads per base pair was normalized to total reads per sample and expressed as a log2 value. Normalized reads per base pair from DNA precipitated from cells expressing only the HA epitope were used as background and subtracted from the other samples. The 62 transcription units were selected on the basis of high signal and isolation from surrounding transcription units. (B) Profiles for the rrnA operon (complement of M. smegmatis 3819731–3825749 with 3825475 set as 0). ORFs are denoted by arrows underneath. Traces are colored as in A.