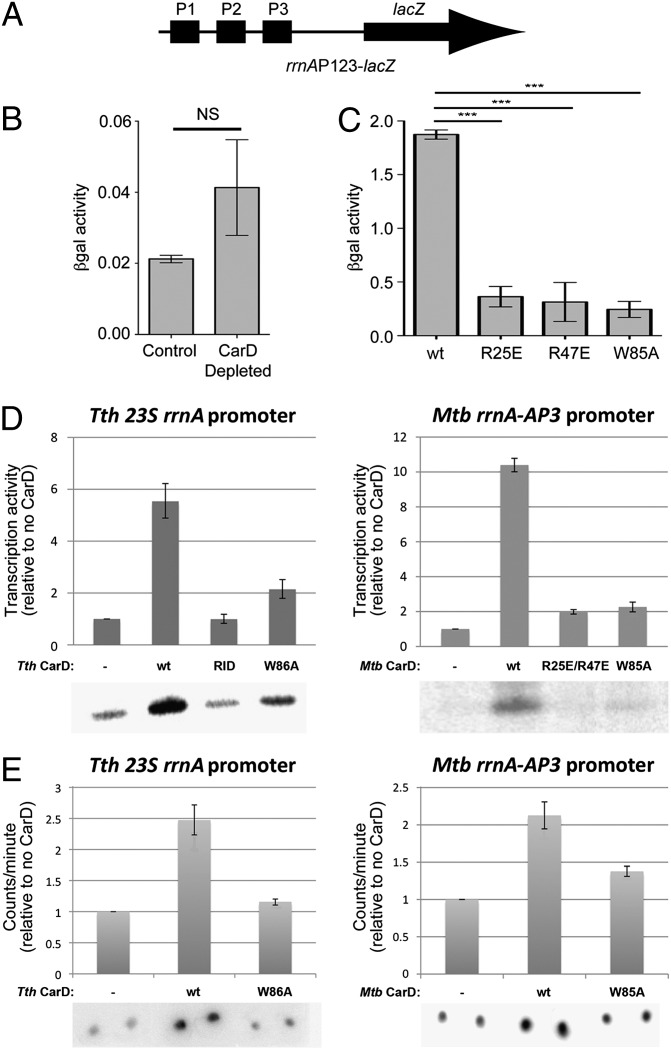
Fig. 4.
CarD activates transcription initiation at rRNA promoters. (A) Schematic illustrating the M. smegmatis rrnAP123-lacZ cassette used for B and C. (B and C) Normalized βgal units of activity in M. smegmatis containing rrnAP123-lacZ (A). The mean of four to five replicates is graphed and the error bars represent the SEM. NS, not significant; ***P value ≤ 0.005 in t tests. (B) Depletion of CarD (1) resulted in increased (approximately twofold) βgal activity. (C) Disruption of the CarD/RNAP protein/protein interaction (R25E or R47E substitutions) (4), as well as the W85A substitution, resulted in decreased (more than fourfold in each case) βgal activity. (D) CarD stimulates transcription in vitro (Left, Thermus transcription system; Right, Mycobacterial transcription system). In each panel, transcription activity (determined by phosphorimagery of 32P-labeled transcripts from denaturing polyacrylamide gels such as those shown below), normalized to no CarD (−), is shown as a histogram. “RID” denotes CarD-RID only (T. thermophilus CarD residues 1–64). The error bars represent the SEM of three replicates. (E) CarD stimulates RPo formation, as determined using filter binding (Left, Thermus transcription system; Right, Mycobacterial transcription system). RNAP and 32P-labeled promoter DNA were incubated to form RPo, challenged with a large excess of unlabeled competitor DNA, and then washed through a nitrocellulose filter to remove unbound DNAs. Bound DNA was quantitated by phosphorimagery. In each panel, binding activity, normalized to no CarD (−), is shown as a histogram. The error bars represent the SEM of four replicates.