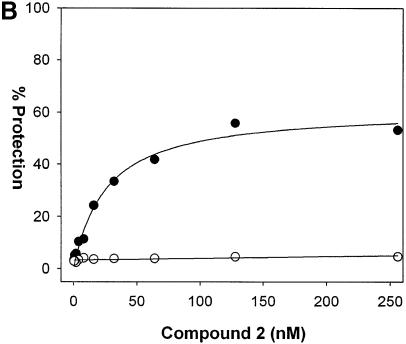
Figure 8.
Binding affinity determination of compound 2. (A) Structure of compound 2. (B) A template DNA containing the sequence TAACAAT·AUUGTTA (match site) in place of the degenerate N(9) sequence was incubated with increasing concentrations of compound 2 and then digested with UDG for 1 min. The extent of protection from UDG was determined by TaqMan PCR analysis as described in Materials and Methods. As a negative control, an oligonucleotide containing a mismatch site TAATAAT·AUUATTA was processed in parallel. Closed circles represent data for the match oligonucleotide and open circles represent data for the mismatch oligonucleotide. The Kd of compound 2 for the match site is determined by fitting the data using Sigma Plot to the equation %Protection = (a × [compound])/(b + [compound]) where a is the maximum %Protection and b is Kd.