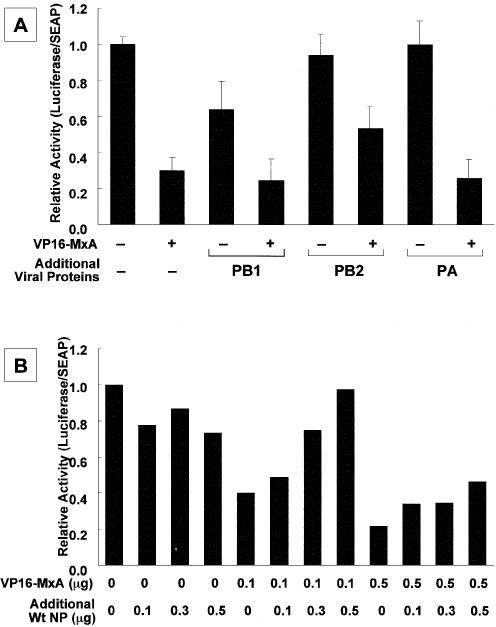
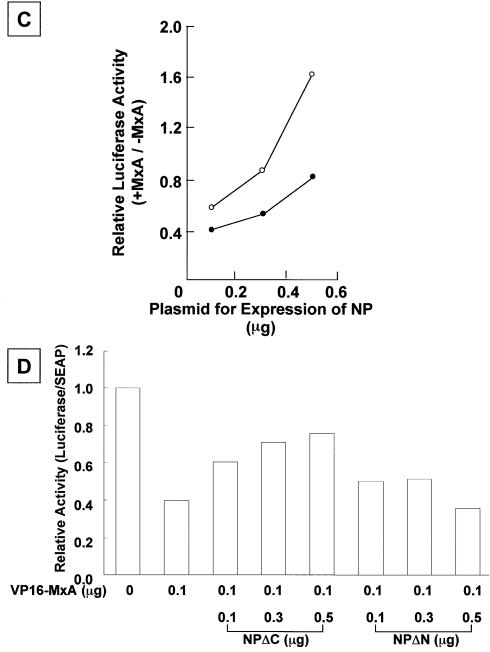
Figure 4.
Rescue of the inhibitory activity of MxA proteins by over- expression of influenza RNA polymerase subunits and NP. (A) Effect of over-expression of viral RNA polymerase subunits. Swiss3T3 cells were transfected with pHMP1-vNS-Luc (0.2 µg), pSEAP (0.1 µg), pVP16-MxA (0 or 0.1 µg indicated – or +, respectively), and three RNA polymerase subunits and NP (0.05 µg each). The additional amount (0.3 µg) of a plasmid encoding either PB1, PB2 or PA was added as indicated. The luciferase activity was determined and shown as described in Materials and Methods. Error bars represent standard deviation (n = 3). (B) Effect of over-expression of NP. Swiss3T3 cells were transfected with pHMP1-vNS-Luc (0.2 µg), pSEAP (0.1 µg), pVP16-MxA (0, 0.1 or 0.5 µg), and three RNA polymerase subunits (0.05 µg each) and NP (0.15 µg). The additional amounts of the plasmid encoding NP were added as indicated. The luciferase activity was determined and shown as described in Materials and Methods. The result is shown as the average of two independent experiments. (C) Titration of plasmid encoding NP. Swiss3T3 cells were transfected with pHMP1-vNS-Luc (0.2 µg), pSEAP (0.1 µg), three RNA polymerase subunits (0.05 µg each) and NP (0.1, 0.3 or 0.5 µg) in the absence or presence of pVP16-MxA. The relative luciferase activity in the presence of MxA (0.1 µg, open circle; 0.3 µg, closed circle) was normalized with that in the absence of MxA, and plotted as a function of increasing amounts of NP. The result is shown as the average of two independent experiments. (D) Titration of VP16-MxA, and C-terminal (NPΔC) and N-terminal (NPΔN) deletion mutants of NP. The same procedure for (B) was carried out with mutant NPs. For expression of NP proteins lacking its C-terminal region (NPΔC) corresponding amino acid positions between 182 and 498 (where amino acid position 1 is set the first amino acid of the wild-type NP) and its N-terminal region (NPΔN) corresponding amino acid positions between 1 and 190, pCAGGS-NPΔC and pCAGGS-NPΔN plasmids were constructed as follows: portions of NP fragments were amplified by PCR with pCAGGS-NP as a template and a set of primers, 5′-TTGAATTCGCCACCATGGCGACCAAAGGCACC-3′ and 5′-TTGAATT CTTAAGCACCTGCGGCCCCAGACC-3′ for NPΔC and a set of primers, 5′-TTGAATTCGCCACCATGGAATTGATCAGAATG-3′ and 5′-GGGA ATTCTTAATTGTCGTACTCCTCTGCA-3′ for NPΔN. Synthesized NP fragments were digested with EcoRI and cloned into pCHA that had been digested with the same restriction enzyme. The additional amounts of the plasmid encoding deletion mutants of NP were transfected as indicated. The result is shown as the average of two independent experiments.