Figure 1.
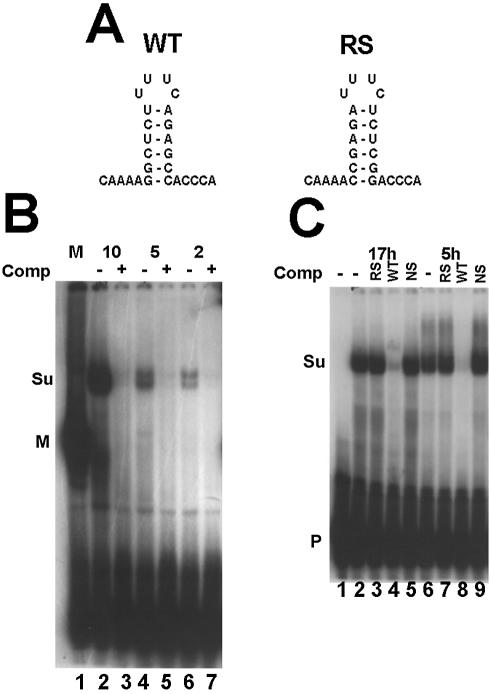
Detection of suSLBP using a mobility shift assay. (A) The sequence of the 3′ end of histone mRNA used in the mobility shift assay is shown as well as the reverse-stem RNA used as a non-specific competitor. (B) An extract (10 mg/ml) was prepared from sea urchin eggs as described in Materials and Methods. Varying amounts (indicated above each lane) of the extract were incubated with the radiolabeled stem–loop probe. In lanes 3, 5 and 7, a 100-fold excess of unlabeled stem–loop (SL) was mixed with the probe prior to addition of the extract. In lane 1, 10 µg of extract from mouse myeloma cell nuclei was used. The complexes were resolved by native gel electrophoresis in a 10% polyacrylamide gel and detected by autoradiography. M is the complex with mouse SLBP and Su the complex with sea urchin SLBP. P indicates the unbound probe. (C) Extracts were prepared from 5 h embryos, or from purified nuclei from 17 h embryos. The extracts were incubated with the radiolabeled stem–loop (lanes 2 and 6). A 100-fold excess of the reverse-stem RNA (RS, lanes 3 and 7), the stem–loop RNA (SL, lanes 4 and 8) or a non-specific oligoribonucleotide (lanes 4 and 9) was added to the probe before addition of the extract. Lane 1 is the probe incubated in buffer. The complexes were resolved as in (B).