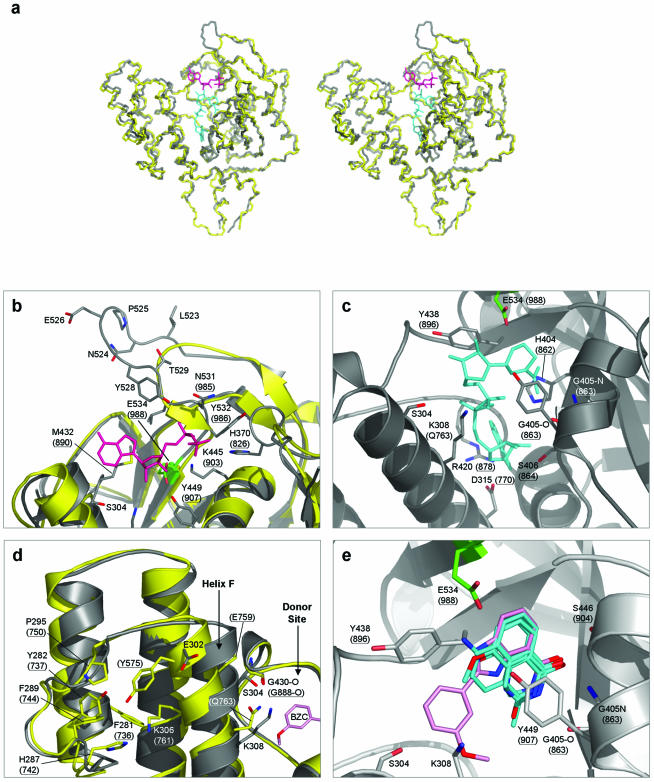
Figure 5.
Amino acid residues shown both in parentheses and underlined are in PARP-1. All other residues are in PARP-2. In addition, cartoon representations of PARP-1 are coloured yellow, and those of PARP-2 coloured grey. The catalytic residue E534 (E988 in PARP-1) is highlighted in green, in all cases. (a) Superimposed Cα traces of PARP-1-CF and PARP-2-CF. Stereo-pair diagram showing superimposed Cα backbone traces for PARP-1-CF (PDB: 2PAW) and PARP-2-CF. Also shown are stick representations of the ADP-ribose moiety of carba-NAD (shown in magenta) and NAD+ (cyan). The positions of carba-NAD and NAD+ were modelled by comparisons with the ligand-bound crystal structures of PARP-1 (PDB: 1A26), and diphtheria toxin (PDB: 1TOX), respectively. (b) Polymer acceptor site. The ADP-ribose moiety of NAD+ (coloured magenta) is shown bound to the donor site of PARP-1 (PDB: 1EFY), along with a superimposition of the equivalent secondary structure elements in PARP-2. All residues involved in coordinating the substrate are highly conserved between the two PARP molecules. The extended loop unique to PARP-2 is clearly visible, consisting of residues Leu523–Thr529. In particular, residue Tyr528 is presented towards the acceptor site, possibly providing additional interactions with the pyrophosphate backbone of the bound substrate. (c) Polymer donor site. A model for NAD+ (shown in cyan) bound to the donor site of PARP-2 is shown. All residues involved in coordination of the substrate are again strongly conserved between PARP-1 and PARP-2, apart from residue Gln763, which is replaced by Lys308 in PARP-2. (d) Helix F in PARP-2 is displaced towards the donor site. In comparison to PARP-1, the top of helix F in PARP-2 is displaced towards the donor site of the molecule. This is due to the replacement of the side-chain of Tyr575 in PARP-1, normally buried in a hydrophobic pocket, with the solvent-exposed residue Glu302 in PARP-2. (e) PARP inhibitors bound to the donor site. PARP inhibitors (for which the structures of complexes with PARP-1 are available) are shown bound to the donor site of PARP-2. Superimposition of topologically equivalent residues, between PARP-1 and PARP-2, facilitated the modelling of these inhibitors. The inhibitor 2-(3′-methoxyphenyl) benzimidazole-4-carboxamide (BZC) is highlighted (shown in pink), demonstrating the proximity of helix F and hence residues Ser304 and Lys308 to the donor site, providing possible targets for future PARP-2 selective/discriminative inhibitors. The inhibitors shown are: 3,4-dihydro-5-methyl-isoquinolinone (DHQ, PDB: 1PAX), 4-amino-1,8-naphthalimide (4AN, PDB: 2PAX), 3-methoxybenzamide (3 MB, PDB: 3PAX) and BZC (PDB: 1EFY).