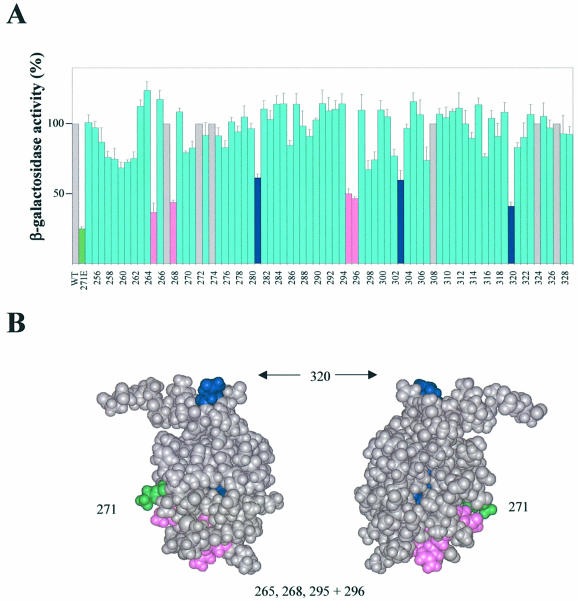
Figure 2.
Identification of αCTD residues important for CII-dependent activation of pE in vivo. (A) Strain WAM105, containing plasmids pTJSpE, pJMH1 and pMO23, was transformed with one of a set of plasmids encoding the RNAP α subunit in which each residue of αCTD (positions 255–329) was changed individually to alanine. Cultures were grown in LB medium containing appropriate antibiotics at 37°C until OD ∼0.2, whereupon IPTG was added to a final concentration of 0.1 mM. After 75 min induction, the β-galactosidase activities were determined. The activities are presented relative to the activity of the strain harbouring pLAW2, encoding wild-type (WT) α (100% = 18 293 Miller units), and are averages of at least three independent experiments. The green bar shows the activity for the strain harbouring pLAW2phs (specifying α K271E). Blue bars and pink bars indicate positions within αCTD at which alanine substitution causes a decrease in activity of ≥40% compared with wild-type α, with the pink bars corresponding to residues within the DNA-binding determinant (7,8). Grey bars indicate positions where alanine occurs naturally (i.e. same as WT α). (B) Structure of αCTD (37), showing amino acid residues that are important for CII-dependent activation of pE. Residues comprising part of the DNA-binding determinant are shown in magenta; other residues are shown in blue (residues 281 and 303 are buried within the αCTD structure and so are not visible in this representation). Lys271 is also highlighted (green).