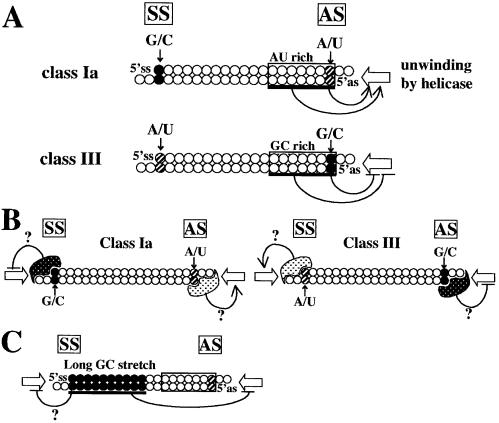
Figure 8.
Possible models of siRNA-based RNAi in mammalian cells. The rules for siRNA sequence preference are schematically shown in (A). siRNA unwinding might be effectively initiated from the AU-rich AS end in the case of class Ia siRNA, lacking a long GC stretch. On the other hand, siRNA duplex unwinding might be suppressed from the GC-rich class III AS end. G/C at the 5′ SS end of class Ia and the 5′ AS end of class III siRNAs might provide a site for binding of an unidentified protein possibly suppressing siRNA unwinding (B). Alternatively, A/U at the 5′ SS end of class III and the 5′ AS end of class Ia siRNAs might serve as a binding site for putative unwinding stimulation factors other than helicase. A long GC stretch such as that found in siRNA n (see Fig. 4B) might prevent the elongation of siRNA duplex denaturation from the AS end (C). A/U at the 5′ AS and SS ends and their counterparts in the SS and AS, respectively, are shown as hatched circles; G or C, closed circles. Terminal AU-rich or GC-rich regions are boxed. Open arrows indicate the direction of siRNA unwinding due to a hypothetical siRNA helicase.